| Welcome, Guest |
You have to register before you can post on our site.
|
| Online Users |
There are currently 64 online users.
» 19 Member(s) | 42 Guest(s)
Bing, Google, Yandex, bernard, Bisou, DeParis, GHurier, jamtastic, old europe, Orentil, PQRS, Qrts, Radko, RBHeadge, Riverman, Rober_tce, theplayer, Vinitharya
|
| Latest Threads |
E-V22 origins and spread
Forum: E
Last Post: Southpaw
6 minutes ago
» Replies: 26
» Views: 612
|
Did the Y-Adam ventured o...
Forum: Ancient (aDNA)
Last Post: GHurier
7 minutes ago
» Replies: 0
» Views: 5
|
Ancient Egyptians
Forum: Ancient (aDNA)
Last Post: Qrts
10 minutes ago
» Replies: 34
» Views: 2,945
|
Ancient DNA Reveals the F...
Forum: Ancient (aDNA)
Last Post: GHurier
28 minutes ago
» Replies: 51
» Views: 2,680
|
Legends
Forum: Genealogy
Last Post: Rufus191
1 hour ago
» Replies: 17
» Views: 756
|
Coming Soon: Y-DNA Haplog...
Forum: FTDNA
Last Post: Mabrams
1 hour ago
» Replies: 333
» Views: 15,157
|
R1b-L51 in Yamnaya: Lazar...
Forum: R1b-L51
Last Post: alanarchae
2 hours ago
» Replies: 353
» Views: 12,262
|
Stolarek et al: Genetic h...
Forum: Ancient (aDNA)
Last Post: ambron
2 hours ago
» Replies: 857
» Views: 66,415
|
Just for fun. What are y...
Forum: Autosomal (auDNA)
Last Post: mjaguk
3 hours ago
» Replies: 49
» Views: 1,964
|
What is methode test mtDN...
Forum: Other
Last Post: Capsian20
7 hours ago
» Replies: 0
» Views: 13
|
The increase of Iran/Zagr...
Forum: Ancient (aDNA)
Last Post: ilabv
9 hours ago
» Replies: 13
» Views: 544
|
Trees
Forum: Fauna & Flora
Last Post: lg16
Yesterday, 11:25 PM
» Replies: 0
» Views: 14
|
Maps of ancient DNA sampl...
Forum: Ancient (aDNA)
Last Post: Anthrofennica
Yesterday, 09:48 PM
» Replies: 1
» Views: 131
|
Shared Roots Matches: Anc...
Forum: African
Last Post: szin
Yesterday, 09:26 PM
» Replies: 1
» Views: 44
|
If there are studies with...
Forum: Ancient (aDNA)
Last Post: Anzor
Yesterday, 09:21 PM
» Replies: 296
» Views: 16,272
|
Basal Eurasian discussion
Forum: Ancient (aDNA)
Last Post: Kale
Yesterday, 09:02 PM
» Replies: 244
» Views: 16,002
|
Right populations to use ...
Forum: Inquiries Corner
Last Post: Kale
Yesterday, 08:59 PM
» Replies: 3
» Views: 57
|
The Genetic Origin of the...
Forum: Ancient (aDNA)
Last Post: Jaska
Yesterday, 08:03 PM
» Replies: 207
» Views: 15,885
|
Bronze Age calculator 202...
Forum: Autosomal (auDNA)
Last Post: Alexander87
Yesterday, 05:38 PM
» Replies: 33
» Views: 1,278
|
Lazaridis 2024 - Associat...
Forum: Natural Sciences
Last Post: Арсен
Yesterday, 05:08 PM
» Replies: 5
» Views: 96
|
old versions of Ancestry ...
Forum: 23andMe
Last Post: szin
Yesterday, 04:19 PM
» Replies: 7
» Views: 411
|
Modern Japanese people ar...
Forum: Ancient (aDNA)
Last Post: qijia
Yesterday, 03:39 PM
» Replies: 9
» Views: 793
|
Datasets for North Africa...
Forum: Inquiries Corner
Last Post: TanTin
Yesterday, 03:16 PM
» Replies: 2
» Views: 39
|
Steppe Ancestry in wester...
Forum: Ancient (aDNA)
Last Post: Strider99
Yesterday, 12:53 PM
» Replies: 398
» Views: 35,024
|
Native Americans from the...
Forum: Autosomal (auDNA)
Last Post: ph2ter
Yesterday, 12:47 PM
» Replies: 285
» Views: 10,706
|
Souk Gaulois
Forum: Western
Last Post: dalluin
Yesterday, 12:07 PM
» Replies: 7
» Views: 307
|
E-V13 - Theories on its O...
Forum: E1b1b-M215
Last Post: Riverman
Yesterday, 10:58 AM
» Replies: 833
» Views: 34,305
|
Dataset merging help
Forum: Inquiries Corner
Last Post: Genetics189291
Yesterday, 09:53 AM
» Replies: 12
» Views: 202
|
R1b-L389: Eastern Branch ...
Forum: R1b general
Last Post: targaryen
Yesterday, 06:24 AM
» Replies: 17
» Views: 729
|
Post your ftdna results
Forum: FTDNA
Last Post: Tolan
Yesterday, 05:38 AM
» Replies: 20
» Views: 922
|
|
|
| Did the Y-Adam ventured outside Africa before coming back and expanding again ? |
|
Posted by: GHurier - 7 minutes ago - Forum: Ancient (aDNA)
- No Replies
|
 |
(I don't think this interpretation of published data have already been proposed, if someone already raised these elements, please feel free to add a link toward the related discussion).
I think most people here are knowing very well the paper from Petr et al. 2020 (https://www.science.org/doi/10.1126/science.abb6460) that unveiled modern human Y-chr as more closely related to Neanderthal Y-chr and more distant to Denisovan Y-chr. Similar results were found for mtDNA (see e.g., Posth et al. 2017, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5500885/).
These results were in tension with autosomal DNA results showing that Denisovans and Neanderthalensis were sharing a common ancestor that already splitted from the Sapiens lineage.
The sequencing of the mtDNA of the Sima de los Huesos Neanderthal sample that unfolded as being more closely related to Denisovan than to modern Human led to the conclusion that Sapiens mtDNA came to replace Neanderthal mtDNA at some point after ~400 kyr BP.
In their paper, Petr et al. 2020 proposed that the same thing occured for Y-DNA, around the same epoch ~370 kyr BP.
However, there is no known Y-DNA from Neanderthals or Sapiens before the claimed replacement. Which in fact opens another possibility, what if it was the other way around and Sapiens Y-chr came to be replaced by a Neanderthal Y-chr that would became ultimately our Y-Adam ?
When we look closely at the results of Posth at al 2017 and Petr et al. 2020 we can notice that :
-Denisovans mtDNA separated from Sapiens mtDNA 1065 +/- 172 kyr ago (1 sigma interval, 68% confidence interval).
-Neanderthal mtDNA separated from Denisovan mtDNA ~785 +/- 137 kyr ago.
-The Sapiens related mtDNA that replaced Neanderthal mtDNA splitted from modern lineage ~414 +/- 27 kyr ago.
Whereas,
-Denisova Y-chr splitted ~707 +/- 50 kyr ago from the shared Y-chr of Sapiens and Neanderthals
-Sapiens and Neanderthal Y-chr splitted ~360 +/- 20 kyr ago.
The main issue with the results of Posth et al. 2017 is the large gap between mtDNA Neanderthal/Denisovan split and their proposed dating of the nDNA split between Neanderthal and Denisovan (~427 +/- 23 kyr ago).
Indeed, for this to work a ~358 +/- 140 kyr old diversity (~2.5 sigma tension with a ~0 kyr diversity hypothesis) for the mtDNA should have been carried by the "rather small" Neandersovan population when they splitted.
The same applied to the source population of Sapiens and Neandersovans split estimation from nDNA (~657 +/- 53 kyr ago), that should have conserved ~400 \pm 180 kyr old mtDNA diversity when they splitted (a ~2.2 sigma tension with a ~0 kyr diversity hypothesis).
By comparison, modern humans, despite being highly diffusive and spatially isolated, failed to conserve anything older than ~150kyr diversity for mtDNA up to modern days.
It is therefore hard to justify ~400 kyr diversity carriage by early human populations. Which cast doubts about the nDNA-based estimation of divergence times between Sapiens, Neanderthals, and Denisovans proposed in Posth et al. 2017 analysis. In particular, all sampled Neanderthals and Denisovans are exhibiting fairly short diversity "horizon", which disfavor the proposition of Neandersovan population carrying very old diversity since their root.
Let consider few hypotheses :
1) mtDNA, Y-chr, and autosomal separation between this three groups (Sapiens, Denisovan, Neanderthal) occurred from small populations carrying few ~kyr old diversity.
2) The events that produced the late mix-up between Sapiens and Neanderthal for mtDNA and Y-chr is a single admixture event.
3) We assume that Posth et al. 2017 mtDNA molecular-clock and Petr et al. 2020 Y-chr molecular clock are consistent.
Under these three hypotheses :
--> The dating from Posth et al. 2017 of the mtDNA splits favors the idea that Sapiens and Neandersovans splitted ~1065 +/- 172 kyr ago, and that Neanderthals and Denisovans splitted ~785 +/- 137 kyr ago. It is interesting to note that such early splitting times for Neanderthal and Denisovan are also proposed by Roger et al. 2020 (https://www.science.org/doi/10.1126/sciadv.aay5483) based on nDNA.
--> We can note a ~390 kyr split between Sapiens and late-Neanderthals mtDNA and Y-chr (assumed to derive from a single event in our hypothesis-2, that support similar molecular clocks calibration from Posth et al. 2017 and Petr et al. 2020), we however note that the two splits are separated by 54 +/- 33 kyr (at ~1.6 sigma from a simultaneous hypothesis).
--> The dating of the Y-Chr split between Sapiens and Denosivans in Petr et al. 2020 (~707 +/- 50) favors the idea that what we are seeing here is not the Sapiens/Neandersovans split (1065 +/- 172 kyr, ~2 sigma tension) but the Neanderthal/Denisovan split (785 +/- 137, ~0.5 sigma agreement). Which would imply that this Y-Chr originally transited by Neanderthal-population before introgressing into Sapiens population.
--> Sometimes around ~360 +/- 20 kyr ago Neanderthals and Sapiens came into contact and mtDNA and Y-chr got exchanged, Sapiens mtDNA introgressed into Neanderthal population and Neanderthal Y-chr introgressed into Sapiens population.
What could kill this interpretation ?
We have seen above that removing hypothesis (1) would cost a lot about the amount of diversity to be carried by the source populations of Sapiens, Neanderthals, and Denisovans. Therefore, I would considered hypothesis (1) as fairly secured grounds.
If we remove hypothesis (2), then we didn't need anymore to have Sapiens/late-Neanderthals split to occur at roughly the same time for mtDNA and Y-chr ... therefore, we can challenge the molecular clocks of Petr et al. 2020 (or Posth et al. 2017) to reconcile the Denisovan/Sapiens split from Y-chr with the same split from mtDNA.
However, removing hypothesis (2) would imply two independent introgression events from Sapiens to Neanderthals that would have ended in mtDNA replacement in one occurence and Y-chr replacement for the other.
Hypothesis (3) is supported by the similar time of mtDNA and Y-chr splits for Sapiens and late-Neanderthals, therefore a miscalibration of the molecular clocks can only accounts for a ~50 kyr mismatch around ~400 kyr between the two datation systems, that remains small (at the order of ~0.5 sigma for the Sapiens/Denisovans split timing).
How could have happened such haplogroup-exchange ?
Here is a proposed scenario:
*A population from ~Sapiens-branch left the Great Rift Valley around ~414 +/- 27 kyr ago, splitting the mtDNA that will be introgressed into Neanderthals from the mtDNA line that will become Mitocondrial-Eve.
* Around ~360 +/- 20 kyr ago this population came into contact with a Neanderthal population. A minimal interaction between the two populations led to the introgression of Sapiens-related mtDNA into Neanderthal population and early Neanderthal Y-chr into the Sapiens-related population.
* These two populations later diffused the introgressed mtDNA (for Neanderthals) and Y-chr (for the Sapiens-related).
In this model, carriers of this Y-chr originating from Neanderthal population before 400 kyr BP would became our Y-Adam around ~236 kyr ago.
The main question that remains open under this model is about "where" to make these two populations interact ?
There is two main realistic options :
1) Around Western Asia
2) Gibraltar Straight (the low-likelyhood of crossing the straight would explain why no significant autosomal DNA signal was exchanged)
As of today, under such model, I would find Gibraltar as the best option. We know that by ~300 kyr modern-humans were around in Morocco at Djebel Irhoud.
In particular, 315 +/- 34 kyr Lellavois-technique tools are documented from this archeological site.
Also the diffusion of Acheulean tools might support the idea of an original Gibraltar-crossing ~700 kyr ago due to the lack of early-Acheulean tools in South-Eastern part of Europe. However, a fast coastal migration from Western Asia toward Iberian peninsula can not be completely ruled out.
The later point is important, as these claimed "contacts" and introgression events can be supported by potential cultural elements diffusion such as :
-Acheulean tools, who reached Europe fairly lately (~700 kyr BP) likely with Homo-Heidelbergensis when the Denisovans/Neanderthal split occurred. Which therefore favors again a high splitting time for Neanderthal and Denisovan around ~700 kyr ago (and thus a required higher dating of the Sapiens/Denisovans split, hard to reconcile with the low-datation of Modern-Sapiens/Denisovans Y-chr from Petr et al. 2020).
-The Levallois tecnique tools, that appears in Africa, Europe and Asia around MIS9 (~330 kyr). That might have been spread by the wave of humans responsible for the mtDNA and Y-chr exchange event. According to Moncel et al. 2020 (https://www.sciencedirect.com/science/ar...via%3Dihub), Levallois Technique tools elaboration is documented from MIS12 to MIS9 in Western Europe.
Therefore, we can propose that this technology spread from South-Western Europe and then the introgression event led to the a back diffusion of this technic in North Western Africa.
Could have the Y-chr of modern human transited among another human subgroup instead of Neanderthal ?
Such scenario would involve a ghost population, probably in North Africa, that was carrying the modern Y-chr line, and that would have splitted from Denisovan around ~700 kyr ago.
Around ~400 kyr ago, this population would have been overwhelmed by Sapiens demography (when Sapiens mtDNA diffused) but got its Y-chr becoming dominent inside the predominantly Sapiens population and introgressing into Neanderthal population.
Yet, traces of archaic population into modern humans genome mostly propose an extra archaic population who splitted away before Sapien/Neanderthal-split, therefore not fitting the needed hierarchy of having a closer relation with Neandersovans than Sapiens.
If such ghost population existed, we have yet to find genetic traces of its existence.
Conclusion :
As we have seen, mtDNA and cultural contacts are favoring an early dating for splits of the populations that would become Sapiens, Neanderthal, and Denisovans (~1Myr for Sapiens/Neandersovans split and ~700kyr for Neanderthals/Denisovans split).
Such early splits are hard to reconcile with the modern-Human/Denisovan Y-chr split that post date the said split by ~360 +/- 180 kyr,
Whereas it better corresponds (with a likelihood ratio of ~10) to the Neanderthal/Denisovan mtDNA split ~785 +/- 137 kyr ago.
The rapid diffusion around the world of Levallois-technique tools around ~330 kyr BP appears to be nearly concomitant with the MRCA of Modern-Humans and late-Neanderthal mtDNA and Y-chr, opening the possibility that the diffusion of this technology might, at least partially, explain the success of the associated lineages.
Interestingly, a careful analysis of the split dating might indicates that Y-Adam ancestor ventured outside Africa before coming back and diffusing again (inside and outside Africa with the ~50 kyr OoA event).
The ideal method to test such possibility would be to have access to a ~450 kyr old Y-chr from a Neanderthal individual (or a Sapiens, but considering sample conservation, this seems less likely to occur).
|

|
|
| Trees |
|
Posted by: lg16 - Yesterday, 11:25 PM - Forum: Fauna & Flora
- No Replies
|
 |
Trees are wonderfull. They do so much for us and this planet. A thread dedicated for all things trees and yes you can call me a tree hugger.
|

|
|
| Lazaridis 2024 - Associated Climate Data |
|
Posted by: Cejo - Yesterday, 01:56 AM - Forum: Natural Sciences
- Replies (5)
|
 |
I wanted to post some climate data for the area and time-frame highlighted in the Lazaridis paper, basically based on this map:
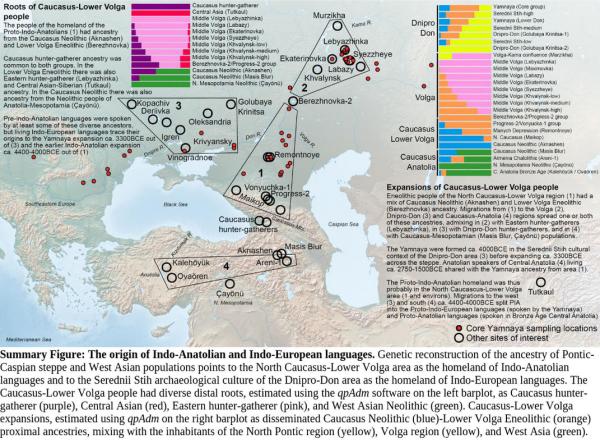
First up are some measurements taken from peat samples, near the southern extent of area 1. These charts show a very significant increase in temperature and a decrease in precipitation between 5500-5000 ybp. The coldest months notably go above freezing, while the warmest months spike to 2x the long-term average, since then. Precipitation drops by 40-50%. Unfortunately, measurements only cover back to 5500 ybp, or 3550 BCE, while the Lazaridis paper indicates migration from this region ca. 4400-4000 BCE (6350-5950 ybp).
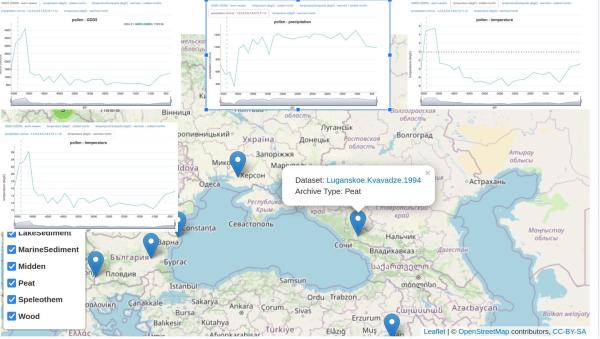
However, I also made some plots from climate models/simulations, which fortuitously focus on 6000 ybp, the period when migration from 1 is indicated. These show 100-year average temperatures for summer and winter months. These show winter temperatures in area 1 generally at or above freezing south, decreasing north. Summer temperatures are warm, in the 23-25 range. Combined with the peat samples above, this indicates that there was a sharp dip in temperatures between 6000 ybp and 5500 ybp, before temperatures spiked again.
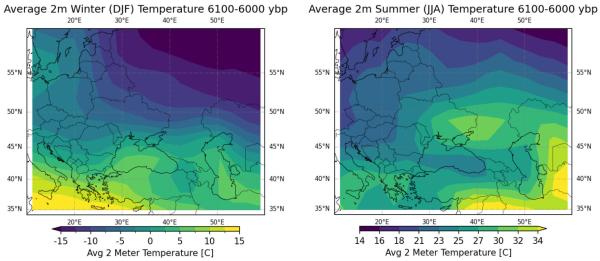
Next up is a look at sediment samples from Lake Van, near area 4 from the study. These temperatures and precipitation plots are a little more stable. While there are oscillations, the magnitude is generally smaller. Potentially worth noting is that precipitation increased roughly 33% prior to indicated migration into the region, and while it oscillated from there, the long term average remained generally elevated for the next 1000 years, dropping again around the time of migration from area 3.
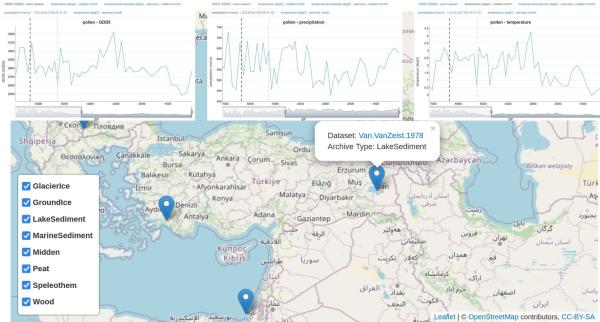
Finally, I have included precipitation and temperature plots from peat samples near area 3. To me, the only really notable change is prior to migration from 1 to 3 (shown in black dotted line), specifically regarding coldest-month temperatures. The indicated date of migration immediately follows recovery from a cold period, during which winters dropped below freezing, returning to above-freezing temperatures year-round. There is a similar reduction and recovery in GDD5 (growing degree-days). I don't notice any massive changes on the later date (grey dotted line) which corresponds to migration from area 3 and expansion across the Steppe.
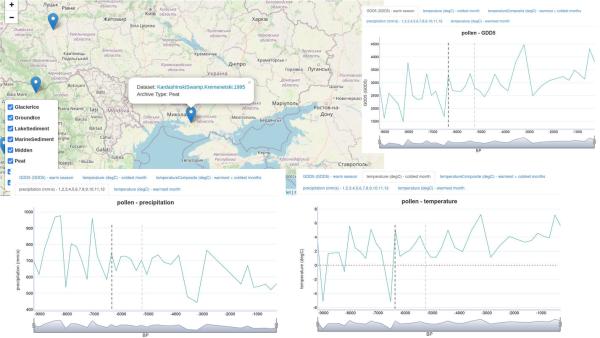
I'm still working on conclusions. It is hard to identify a climate-driven motivation for migrations from area 1, given discontinuity in data, aside from a hint of some temperature changes. However, one general pattern that emerges is expansion into areas recovering from changes. In other words, it doesn't seem to have been necessarily local climate changes driving people from an area. Rather, it may have been that nearby areas were put under stress and became relatively depopulated, opening up room for expansion and drawing people to new areas. I think particularly if these areas were inhabited by farmers, and precipitation or temperatures drastically changed, this would have put stress on that kind of economy. This would have been particularly true if plague also accompanied it. None of this is groundbreaking work, but I think that's what I'm seeing here, and seeing this made me think about it a little differently.
Edit: Adding sources...
Oghaito et al 2021 - https://gmd.copernicus.org/articles/14/1195/2021/ (ultimate source of the gridded model climate data I used)
https://esgf-node.ipsl.upmc.fr/search/cmip6-ipsl/ (where I accessed the gridded model climate data)
https://lipdverse.org/Temp12k/current_version/ (where I accessed peat and lake sediment samples)
|

|
|
| Italians |
|
Posted by: HDG33 - 05-06-2024, 05:18 PM - Forum: Southern
- No Replies
|
 |
I created this thread to discuss Italian genetics and its history
|

|
|
|