Posts: 30
Threads: 4
Joined: Oct 2023
Gender: Male
Ethnicity: Saharan
![[Image: sharedroots-oceania.webp]](https://www.dnagenics.com/img/ancestry/sharedroots/sharedroots-oceania.webp)
Find shared connection to ancient samples at chromosomal level
https://www.dnagenics.com/ancestry/share...entoceania
List of samples
Code: EFE005 - Vanuatu
Ancient Individual from Efate, Vanuatu (200 BP) - Located at Banana Bay
FUT001 - Vanuatu
Ancient Individual from Futuna, Vanuatu (1100 BP)
I10967 - Vanuatu
Ancient Polynesian Individual from Efate, Vanuatu (200 BP) - Located at Mangaasi
I10968 - Vanuatu
Ancient Individual from Retoka, Vanuatu (400 BP)
I10969 - Vanuatu
Ancient Individual from Retoka, Vanuatu (400 BP)
I1368 - Vanuatu
Ancient Individual from Efate, Vanuatu (2900 BP) - Located at Teouma
I1369 - Vanuatu
Ancient Individual from Efate, Vanuatu (2900 BP) - Located at Teouma
I1370 - Vanuatu
Ancient Individual from Efate, Vanuatu (2900 BP) - Located at Teouma
I14493 - Vanuatu
Vanuatu 400BP - Retoka
I30476 - Federated States of Micronesia
Individual of Saudeleur Dynasty from Temwen Island, Pohnpei, Caroline Islands, Micronesia - Located at Man Nadol, Pahndauwas
I30477 - Federated States of Micronesia
Individual of Saudeleur Dynasty from Temwen Island, Pohnpei, Caroline Islands, Micronesia - Located at Man Nadol, Pahndauwas
I30478 - Federated States of Micronesia
Individual of Saudeleur Dynasty from Temwen Island, Pohnpei, Caroline Islands, Micronesia - Located at Man Nadol, Pahndauwas
I30480 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
I30481 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
I30482 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
I30483 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
I3921 - Vanuatu
Burumbe Resident of Epi Island, Vanuatu (1300BP)
I4096 - Vanuatu
Burumbe Resident of Epi Island, Vanuatu (1300BP)
I4105 - Vanuatu
Wam Bay Resident of Epi Island, Vanuatu (150BP)
I4424 - Vanuatu
Pangpang Resident of Efate, Vanuatu (150BP)
I4425 - Vanuatu
Ifira Island Resident of Efate, Vanuatu (150BP)
I4450 - Vanuatu
Mele-Taplins Resident of Efate, Vanuatu (2300BP)
I4451 - Vanuatu
Mele-Taplins Resident of Efate, Vanuatu (2300BP)
I5259 - Vanuatu
Mangalitu Resident of Efate, Vanuatu (500BP)
I5265 - Vanuatu
Teouma Resident of Efate, Vanuatu (3000BP)
I6188 - Vanuatu
Mele-Taplins Resident of Efate, Vanuatu (2400BP)
I7111 - Federated States of Micronesia
Pohnpeian Resident of the Caroline Islands, Micronesia (400BP)
LHA001 - Tonga
Tongatapu Resident of Tonga (800BP)
MAI002 - Solomon Islands
Malaitan Resident of the Solomon Islands (500BP)
MAL001 - Vanuatu
Malakulan Resident of Vanuatu (2200BP)
MAL002 - Vanuatu
Malakulan Resident of Vanuatu (2500BP)
MAL006 - Vanuatu
Malakulan Resident of Vanuatu (2700BP)
MAL007 - Vanuatu
Malakulan Resident of Vanuatu (2800BP)
RBC1 - Guam
Resident of the Late Unai Ritidian Phase, Ritidian Site, Guam
Sk10 - Tonga
Resident of Talasiu Site, Tongatapu, Tonga (2700BP)
TAN002 - Vanuatu
Resident of Tanna, Vanuatu (2500BP)
TAP002 - French Polynesia
Resident of Ra'iatea, French Polynesia (200BP)
TON001 - Tonga
Resident of Talasiu Site, Tongatapu, Tonga (2700BP)
Posts: 30
Threads: 4
Joined: Oct 2023
Gender: Male
Ethnicity: Saharan
Me: No matched samples
Dad:
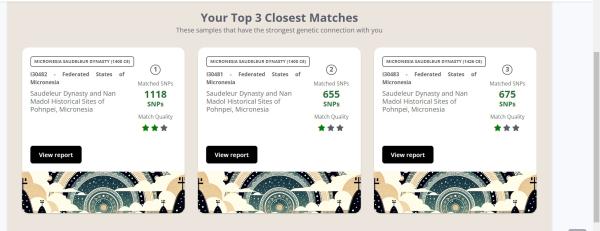
Micronesia Saudeleur Dynasty (1400 CE)
I30482 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
Matched SNPs
213 SNPs
Mom
Match#1 Micronesia Pohnpei 400Bp (1501 CE) - I7111 - Federated States of Micronesia
- Pohnpeian Resident of the Caroline Islands, Micronesia (400BP)
Matched SNPs
518 SNPs (2 segments of 259)
Match#2 Micronesia Saudeleur Dynasty (1409 CE)
- I30480 - Federated States of Micronesia
- Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
Matched SNPs
271 SNPs (1 segment of 271)
Posts: 196
Threads: 33
Joined: Nov 2023
Gender: Male
Ethnicity: Arab
It says I have to pay to upgrade but is everyone that says unlock report related to my son because he scored vanuatu about 2-4 percent in his ancients
a
Posts: 30
Threads: 4
Joined: Oct 2023
Gender: Male
Ethnicity: Saharan
(01-21-2024, 02:27 PM)Genetics189291 Wrote: It says I have to pay to upgrade but is everyone that says unlock report related to my son because he scored vanuatu about 2-4 percent in his ancients
No not everyone.
Posts: 172
Threads: 9
Joined: Oct 2023
Gender: Male
Ethnicity: NWEuro/Maghrebi Andalusi
TOP 3
1
MICRONESIA SAUDELEUR DYNASTY (1644 CE)
I30477 - Federated States of Micronesia
Individual of Saudeleur Dynasty from Temwen Island, Pohnpei, Caroline Islands, Micronesia - Located at Man Nadol, Pahndauwas
Matched 204 SNPs
2
MICRONESIA SAUDELEUR DYNASTY (1400 CE)
I30482 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
Matched 372 SNPs
3
MICRONESIA SAUDELEUR DYNASTY (1400 CE)
I30481 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
Matched 2197 SNPs
Other Matches
These samples show a genetic connection with you
MICRONESIA SAUDELEUR DYNASTY (1409 CE)
I30480 - Federated States of Micronesia
Saudeleur Dynasty and Nan Madol Historical Sites of Pohnpei, Micronesia
Matched 224 SNPs
يَا أَيُّهَا النَّاسُ إِنَّا خَلَقْنَاكُم مِّن ذَكَرٍ وَأُنثَىٰ وَجَعَلْنَاكُمْ شُعُوبًا وَقَبَائِلَ لِتَعَارَفُوا ۚ إِنَّ أَكْرَمَكُمْ عِندَ اللَّهِ أَتْقَاكُمْ ۚ إِنَّ اللَّهَ عَلِيمٌ خَبِيرٌ
S.49:13
Code: Unscaled
0.01794617 Spain_Roman_oMixed
0.01889478 Portugal_MonteDaNora_LateRoman
0.01890273 Austria_Ovilava_Roman
Posts: 610
Threads: 62
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
Actually i dont have any ida about reliability if this true or wrong, but anyway this my shared roots ancient in Ancient Oceania
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 30
Threads: 4
Joined: Oct 2023
Gender: Male
Ethnicity: Saharan
(01-23-2024, 09:24 PM)Capsian20 Wrote: Actually i dont have any ida about reliability if this true or wrong, but anyway this my shared roots ancient in Ancient Oceania
There are more infos on each sample when you click on "view infos".
For instance, for I30482 you can see the autosomal results
![[Image: 8T0HU2L.png]](https://i.imgur.com/8T0HU2L.png)
and have access to the G25 coordinates.
Code: I30482,-0.00481056,-0.37675686,-0.11026404,0.04613044,0.13986554,-0.1030525,-0.00096804,-0.00184282,-0.02349632,-0.01145832,0.00164406,0.0019782,0.00261772,-0.00694184,0.00198646,0.0020435,0.00590472,-0.00314646,-0.00171082,-0.00888026,0.01027376,0.00766192,0.01180692,0.0041648,0.00520136
Target: I30482
Distance: 0.5982% / 0.00598175
63.0 Dai
24.8 Papuan
5.6 Changshan_Yao_Guizhou
5.4 Australian
0.4 Kaba
0.4 Nadar
0.4 She
Posts: 172
Threads: 9
Joined: Oct 2023
Gender: Male
Ethnicity: NWEuro/Maghrebi Andalusi
01-25-2024, 01:44 PM
(This post was last modified: 01-25-2024, 01:48 PM by Mulay 'Abdullah.)
(01-24-2024, 08:05 PM)HurricaneSeason Wrote: (01-23-2024, 09:24 PM)Capsian20 Wrote: Actually i dont have any ida about reliability if this true or wrong, but anyway this my shared roots ancient in Ancient Oceania
There are more infos on each sample when you click on "view infos".
For instance, for I30482 you can see the autosomal results
![[Image: 8T0HU2L.png]](https://i.imgur.com/8T0HU2L.png)
and have access to the G25 coordinates.
Code: I30482,-0.00481056,-0.37675686,-0.11026404,0.04613044,0.13986554,-0.1030525,-0.00096804,-0.00184282,-0.02349632,-0.01145832,0.00164406,0.0019782,0.00261772,-0.00694184,0.00198646,0.0020435,0.00590472,-0.00314646,-0.00171082,-0.00888026,0.01027376,0.00766192,0.01180692,0.0041648,0.00520136
Target: I30482
Distance: 0.5982% / 0.00598175
63.0 Dai
24.8 Papuan
5.6 Changshan_Yao_Guizhou
5.4 Australian
0.4 Kaba
0.4 Nadar
0.4 She
Yeah for that, they make a good job, with nice and useful informations etc, but concerning the point of their coordinates they have the same format than G25, but they are not G25 properly. As example with their coords, I tend to be a little more southern shifted with less northern euro admixture, and more Miiddle Eastern shifted but in other hand less "african shifted" (well in fact I aim especially the north african here). In comparison in this view with the G25 coords my results with coords from DNA Genics seem more weirds.
Globally on a PCA it's not the same but it's near, it's close.
We therefore unfortunately cannot make a real comparison, at least in a serious way, with the G25 coordinates that we have and use with the data sheets that we have from Davidski.
It would be nice if they also offered their own datasheet based on the method they use to produce their coordinates in the G25 format
يَا أَيُّهَا النَّاسُ إِنَّا خَلَقْنَاكُم مِّن ذَكَرٍ وَأُنثَىٰ وَجَعَلْنَاكُمْ شُعُوبًا وَقَبَائِلَ لِتَعَارَفُوا ۚ إِنَّ أَكْرَمَكُمْ عِندَ اللَّهِ أَتْقَاكُمْ ۚ إِنَّ اللَّهَ عَلِيمٌ خَبِيرٌ
S.49:13
Code: Unscaled
0.01794617 Spain_Roman_oMixed
0.01889478 Portugal_MonteDaNora_LateRoman
0.01890273 Austria_Ovilava_Roman
Posts: 172
Threads: 9
Joined: Oct 2023
Gender: Male
Ethnicity: NWEuro/Maghrebi Andalusi
05-03-2024, 01:58 AM
(This post was last modified: 05-03-2024, 02:00 AM by Mulay 'Abdullah.)
My previous results was with Ancestry's data, now with Ancestry I have no matches and yeah I think that makes more sense for me....
They updated the results:
Scientific Details
Algorithm version: 2.4 - April 2024
Reference dataset updated: March 2024
The human population is a complex and branching network of interconnected lineages that started in Africa where they diverged, branched out, and spread across the world. Rather than a simple, direct line of ancestors, it involves numerous branches, splits, and intermixing between different groups of people throughout history. This complexity arises from factors like migration, interbreeding, and the constant diversification of human populations over time. As a result, tracing one's ancestry back in a straight, linear manner becomes increasingly challenging as we go further back in time due to the intricate web of connections among individuals and groups.
Shared Roots Matches report aims to identify small segments of your DNA by comparing each segment with the DNA from ancient samples. The goal is to find the closest and most accurate match possible. To ensure the validity of these matches, specific criteria are employed, which are elaborated upon below.
How does this analysis work?
The comparison analysis of any of these samples with your genome utilizes an Identity-by-State (IBS) approach. This method is particularly effective when dealing with ancient DNA samples, where traditional DNA matching segment analysis methods may not be applicable due to the antiquity. By analyzing small segments that are relevant and genetically unique, the IBS approach allows us to identify shared genetic segments more accurately.
However, it's important to bear in mind that genetic analysis is a field of science that is continually advancing. The current results represent the best outcome achievable with our current knowledge. As the science continues to evolve, some of these results may change, potentially leading to improvements in the accuracy and understanding of the results.
Our algorithm considers multiple factors to determine your connection to this sample:
The number of Single Nucleotide Polymorphisms (SNPs) present in the DNA segment.
The chromosome region in which the segment is located.
The distance between the first and last position of each segment.
The frequency of the segment among the worldwide population, compared to its frequency among populations with similar ancestry of this sample.
The Match Quality Score
The match quality score is a measure of the reliability of the analysis. It is calculated by taking into account the number of SNPs, the length of the segment, and the frequency of the segment among the worldwide population, compared to its frequency among populations with similar ancestry of this sample.
How the quantity of SNPs in the RAW file influences the matching
Depending on the raw data file of your sample, it is possible that your raw file may contain more SNPs per centimorgan. Consequently, the number of shared SNPs may be greater compared to another raw file that matches the same segments.
References
The analysis method employed in this report is well-established within the scientific community. For your reference, we have adopted a similar approach in this analysis to that described in the following scientific papers:
Mapping co-ancestry connections between the genome of a Medieval individual and modern Europeans
Ancient DNA reveals admixture history and endogamy in the prehistoric Aegean
Ancient and recent admixture layers in Sicily and Southern Italy trace multiple migration routes along the Mediterranean
يَا أَيُّهَا النَّاسُ إِنَّا خَلَقْنَاكُم مِّن ذَكَرٍ وَأُنثَىٰ وَجَعَلْنَاكُمْ شُعُوبًا وَقَبَائِلَ لِتَعَارَفُوا ۚ إِنَّ أَكْرَمَكُمْ عِندَ اللَّهِ أَتْقَاكُمْ ۚ إِنَّ اللَّهَ عَلِيمٌ خَبِيرٌ
S.49:13
Code: Unscaled
0.01794617 Spain_Roman_oMixed
0.01889478 Portugal_MonteDaNora_LateRoman
0.01890273 Austria_Ovilava_Roman
Posts: 20
Threads: 2
Joined: Oct 2023
Gender: Male
Ethnicity: Flemish/Serbian
Nationality: French
Y-DNA (P): I-Y6889*
Y-DNA (M): K1a13a2*
Country: 
05-03-2024, 08:10 AM
(This post was last modified: 05-03-2024, 08:11 AM by Square.)
"Oceania Ancient - Vanuatu - Malakula MAL002"
255 total shared SNPs. 4 cm on chr11.
Is this really reliable ? It's seems unlikely to me that I could match such samples.
Dna genics says :
"it does not imply that this sample is your direct ancestor; it simply indicates that both you and this sample share a small piece of DNA that could be related to a common ancestor thousand of years ago."
How I could have a common ancestor, even in the last thousand of years, with people from such geographical distance ?
It would be plausible to me that I can have a match at about 4-6 centimorgans with any European medieval individual from Spain to Belarussia... but ancient Oceanians really ?
Mapping co-ancestry connections between the genome of a Medieval individual and modern Europeans
https://www.nature.com/articles/s41598-020-64007-2
"Generally, European IBD blocks longer than 4 cM derive from common ancestors living 500–1500 years ago or even more recently11, which is a period roughly contemporaneous to our Medieval individual. The network based on >6 cM genomic blocks in modern Europeans uncovers some interesting genealogical features that are not evident in the commonly applied population genetic methods such as PCAs or ADMIXTURE analyses. "
HurricaneSeason likes this post
Posts: 35
Threads: 5
Joined: Apr 2024
Gender: Undisclosed
Ethnicity: Irish, English, Romani etc.
Nationality: United Kingdom (England)
Y-DNA (P): possibly R1b
Y-DNA (M): possibly R-Y90180
mtDNA (M): H1a
Country: 
(05-03-2024, 08:10 AM)Square Wrote: "Oceania Ancient - Vanuatu - Malakula MAL002"
255 total shared SNPs. 4 cm on chr11.
Is this really reliable ? It's seems unlikely to me that I could match such samples.
It could indicate ancient East or South Asian ancestry- not entirely out of the realm of possibility given the Huns and other Steppe populations crossing into Europe.
As an example: I often get tiny amounts of Oceanian/Aboriginal/Andamanese on G25 runs. In my case, I know this to be most likely a signal of an Austronesian population like the Santali who contributed to the Romani gene pool. You're half-Serbian so more likely to have Hungarian ancestors in genetic time, potentially with not insignificant East Asian admix. In my case, you have to go back to the thirteenth century to find a Hungarian ancestor. Despite this ancestor being significantly East Asian admixed (her mother being of the Cumans), I score next to no East Asian on calculators.
Anyway, they updated the algorithm and added new populations. I don't match with as many as I do on the Ancient African Shared Roots, which is surprising:
HurricaneSeason likes this post
Avatar: The obverse of a coin of Kanishka I depicting the Buddha, with the Greco-Bactrian legend ΒΟΔΔΟ.
Follow my attempt at reviving Pictish.
Romanes-lekhipen- the Romani alphabet.
|
![[Image: sharedroots-oceania.webp]](https://www.dnagenics.com/img/ancestry/sharedroots/sharedroots-oceania.webp)






![[Image: 8T0HU2L.png]](https://i.imgur.com/8T0HU2L.png)

