Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
JonikW, lg16, Strider99 like this post
Posts: 6
Threads: 2
Joined: Oct 2023
Gender: Male
Nationality: Spanish
Y-DNA (P): E-FT4968
mtDNA (M): V6b1b
(09-30-2023, 06:36 PM)Capsian20 Wrote: Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
In the two Spanish samples, the subclade is not known?
Posts: 318
Threads: 3
Joined: Sep 2023
(10-21-2023, 09:21 PM)JJJ Wrote: (09-30-2023, 06:36 PM)Capsian20 Wrote: Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
In the two Spanish samples, the subclade is not known? I don't know whether Capsian knows more but in my experience this is all too common with V samples. Even FTDNA still has me at V, but thanks to YFull I know I'm currently at V78. We still today even see studies that only get to HV0 rather than the potential V because they don't look at the coding region.
Capsian20 and JJJ like this post
Y: I1 Z140+ FT354410+; mtDNA: V78
Recent tree: mainly West Country England and Southeast Wales
Y line: Peak District, c.1300. Swedish IA/VA matches; last = 715AD YFull, 849AD FTDNA
mtDNA: Llanvihangel Pont-y-moile, 1825
Mother's Y: R-BY11922+; Llanvair Discoed, 1770
Avatar: Welsh Borders hillfort, 1980s
Anthrogenica member 2015-23
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
(10-21-2023, 09:21 PM)JJJ Wrote: (09-30-2023, 06:36 PM)Capsian20 Wrote: Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
In the two Spanish samples, the subclade is not known?
Yes unknown subclade
JonikW and JJJ like this post
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
(10-21-2023, 09:41 PM)JonikW Wrote: (10-21-2023, 09:21 PM)JJJ Wrote: (09-30-2023, 06:36 PM)Capsian20 Wrote: Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
In the two Spanish samples, the subclade is not known? I don't know whether Capsian knows more but in my experience this is all too common with V samples. Even FTDNA still has me at V, but thanks to YFull I know I'm currently at V78. We still today even see studies that only get to HV0 rather than the potential V because they don't look at the coding region.
you right JonikW , in FTDNA there just 28 subclades so many samples its new subclades ( V29 to 97) on MTree but its still V
My result in 23andME it's still only V
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
My result in 23andME and WGS


JonikW and JJJ like this post
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 6
Threads: 2
Joined: Oct 2023
Gender: Male
Nationality: Spanish
Y-DNA (P): E-FT4968
mtDNA (M): V6b1b
10-22-2023, 09:04 AM
(This post was last modified: 10-22-2023, 09:06 AM by JJJ.)
(10-21-2023, 10:00 PM)Capsian20 Wrote: (10-21-2023, 09:41 PM)JonikW Wrote: (10-21-2023, 09:21 PM)JJJ Wrote: (09-30-2023, 06:36 PM)Capsian20 Wrote: Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
In the two Spanish samples, the subclade is not known? I don't know whether Capsian knows more but in my experience this is all too common with V samples. Even FTDNA still has me at V, but thanks to YFull I know I'm currently at V78. We still today even see studies that only get to HV0 rather than the potential V because they don't look at the coding region.
you right JonikW , in FTDNA there just 28 subclades so many samples its new subclades ( V29 to 97) on MTree but its still V
My result in 23andME it's still only V
They gave me HV and they were satisfied xd
I was told a trick to rule out HV from V by looking at the raw data of 23andme and sure enough I managed to figure out it was V, a WGS test confirmed it and gave me a subclade on Yfull.
![[Image: VmEA29B.jpg]](https://i.imgur.com/VmEA29B.jpg)
Capsian20 and JonikW like this post
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
(10-22-2023, 09:04 AM)JJJ Wrote: (10-21-2023, 10:00 PM)Capsian20 Wrote: (10-21-2023, 09:41 PM)JonikW Wrote: (10-21-2023, 09:21 PM)JJJ Wrote: (09-30-2023, 06:36 PM)Capsian20 Wrote: Hello
Here it is Map ancient Haplogroup V
it will be updated as new sampes get published
https://umap.openstreetmap.fr/fr/map/mtd...3.11/18.25
Unfortunately i need to create new Map because i can't do update this new samples from Poland
PCA0086:V1a1
PCA0113: V
PCA0120: V2c
PCA0130: V1a1
PCA0148:V18a
PCA0206: V7b
PCA0244: V1a1
PCA0246: V1a1
PCA0339:V2c
PCA0483:V3c
PCA488: V3c
PCA0524:V
In the two Spanish samples, the subclade is not known? I don't know whether Capsian knows more but in my experience this is all too common with V samples. Even FTDNA still has me at V, but thanks to YFull I know I'm currently at V78. We still today even see studies that only get to HV0 rather than the potential V because they don't look at the coding region.
you right JonikW , in FTDNA there just 28 subclades so many samples its new subclades ( V29 to 97) on MTree but its still V
My result in 23andME it's still only V
They gave me HV and they were satisfied xd
I was told a trick to rule out HV from V by looking at the raw data of 23andme and sure enough I managed to figure out it was V, a WGS test confirmed it and gave me a subclade on Yfull.
![[Image: VmEA29B.jpg]](https://i.imgur.com/VmEA29B.jpg)
Oh this so interested so maybe people HV0a are also V
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 450
Threads: 6
Joined: Sep 2023
Gender: Male
Ethnicity: Kaikavian Croatian
Nationality: Croatian
Y-DNA (P): I2-A815-FGC92673-Y246976
mtDNA (M): HV0-T195C!-a24
(10-22-2023, 09:31 AM)Capsian20 Wrote: Oh this so interested so maybe people HV0a are also V V is only one of several HV0a branches.
Capsian20 and JonikW like this post
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
(10-22-2023, 09:40 AM)ph2ter Wrote: (10-22-2023, 09:31 AM)Capsian20 Wrote: Oh this so interested so maybe people HV0a are also V V is only one of several HV0a branches.
Yes i know this there HV0a1 , HV0a2 , HV0a3 but i just hope hhh ( I just joking )
have good day ph2tf
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 318
Threads: 3
Joined: Sep 2023
(10-22-2023, 09:31 AM)Capsian20 Wrote: (10-22-2023, 09:04 AM)JJJ Wrote: (10-21-2023, 10:00 PM)Capsian20 Wrote: (10-21-2023, 09:41 PM)JonikW Wrote: (10-21-2023, 09:21 PM)JJJ Wrote: In the two Spanish samples, the subclade is not known? I don't know whether Capsian knows more but in my experience this is all too common with V samples. Even FTDNA still has me at V, but thanks to YFull I know I'm currently at V78. We still today even see studies that only get to HV0 rather than the potential V because they don't look at the coding region.
you right JonikW , in FTDNA there just 28 subclades so many samples its new subclades ( V29 to 97) on MTree but its still V
My result in 23andME it's still only V
They gave me HV and they were satisfied xd
I was told a trick to rule out HV from V by looking at the raw data of 23andme and sure enough I managed to figure out it was V, a WGS test confirmed it and gave me a subclade on Yfull.
![[Image: VmEA29B.jpg]](https://i.imgur.com/VmEA29B.jpg)
Oh this so interested so maybe people HV0a are also V
The odds are certainly in favour of some of the HV0 aDNA samples that have been published over the years in fact being V.
Capsian20 likes this post
Y: I1 Z140+ FT354410+; mtDNA: V78
Recent tree: mainly West Country England and Southeast Wales
Y line: Peak District, c.1300. Swedish IA/VA matches; last = 715AD YFull, 849AD FTDNA
mtDNA: Llanvihangel Pont-y-moile, 1825
Mother's Y: R-BY11922+; Llanvair Discoed, 1770
Avatar: Welsh Borders hillfort, 1980s
Anthrogenica member 2015-23
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
(10-22-2023, 10:09 AM)JonikW Wrote: (10-22-2023, 09:31 AM)Capsian20 Wrote: (10-22-2023, 09:04 AM)JJJ Wrote: (10-21-2023, 10:00 PM)Capsian20 Wrote: (10-21-2023, 09:41 PM)JonikW Wrote: I don't know whether Capsian knows more but in my experience this is all too common with V samples. Even FTDNA still has me at V, but thanks to YFull I know I'm currently at V78. We still today even see studies that only get to HV0 rather than the potential V because they don't look at the coding region.
you right JonikW , in FTDNA there just 28 subclades so many samples its new subclades ( V29 to 97) on MTree but its still V
My result in 23andME it's still only V
They gave me HV and they were satisfied xd
I was told a trick to rule out HV from V by looking at the raw data of 23andme and sure enough I managed to figure out it was V, a WGS test confirmed it and gave me a subclade on Yfull.
![[Image: VmEA29B.jpg]](https://i.imgur.com/VmEA29B.jpg)
Oh this so interested so maybe people HV0a are also V
The odds are certainly in favour of some of the HV0 aDNA samples that have been published over the years in fact being V.
but i think there isn't there any updating in 23andME about subclades mtDNA to yet
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
10-22-2023, 10:36 AM
(This post was last modified: 10-22-2023, 10:36 AM by Capsian20.)
Very intersted for my file 23anndME
I uploded my file to JamesLick and studio DNA kit 2.8 and tellmegen
I got subclade V12
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
New samples from Serbia and Croatia
I15533: V1a1 (200-300 CE) from Serbia
I23304: V (100-300 CE) from Serbia
I26718: V16 (100-400 CE) from Croatia
I27295: V7 (450-550 CE) from Serbia
https://www.cell.com/cell/fulltext/S0092...23)01135-2
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
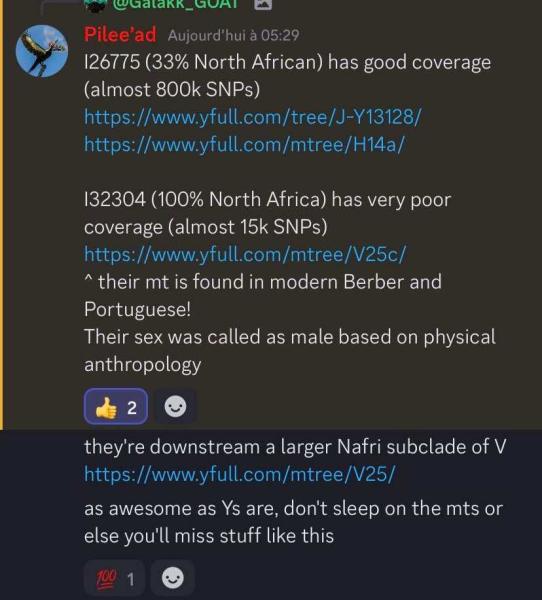
Very interesting my friend send me this picture I32304 it's very possible belong a mtDNA V25 ( V25c ) Iberians and North Africans

Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
|




![[Image: VmEA29B.jpg]](https://i.imgur.com/VmEA29B.jpg)