Posts: 31
Threads: 2
Joined: Nov 2023
Gender: Male
Y-DNA (P): R1b-U152
mtDNA (M): H1
Country: 
Abstract
Background: The Italic Iron Age was characterized by the presence of various ethnic groups partially examined from a genomic perspective. To explore the evolution of Iron Age Italic populations and the genetic impact of Romanization, we focused on the Picenes, one of the most fascinating pre-Roman civilizations, who flourished on the Middle Adriatic side of Central Italy between the 9th and the 3rd century BCE, until the Roman colonization. Results: We analyzed more than 50 samples, spanning more than 1,000 years of history from the Iron Age to Late Antiquity. Despite cultural diversity, our analysis reveals no major differences between the Picenes and other coeval populations, suggesting a shared genetic history of the Central Italian Iron Age ethnic groups. Nevertheless, a slight genetic differentiation between populations along the Adriatic and Tyrrhenian coasts can be observed, possibly due to genetic contacts between populations residing on the Italian and Balkan shores of the Adriatic Sea. Additionally, we found several individuals with ancestries deviating from their general population. Lastly, In the Late Antiquity period, the genetic landscape of the Middle Adriatic region drastically changed, indicating a relevant influx from the Near East. Conclusions: Our findings, consistently with archeological hypotheses, suggest genetic interactions across the Adriatic Sea during the Bronze/Iron Age and a high level of individual mobility typical of cosmopolitan societies. Finally, we highlighted the role of the Roman Empire in shaping genetic and phenotypic changes that greatly impacted the Italian peninsula.
https://www.biorxiv.org/content/10.1101/...8.585512v1
Posts: 710
Threads: 12
Joined: Sep 2023
From the paper:
Quote:Y chromosome data of the Italic IA groups provide additional evidence to these observations,
suggesting that the two scenarios proposed are complementary. Indeed, in the Picenes, two main Y
haplogroups are observed, namely R1-M269/L23 (58% of the total) and J2-M172/M12 (25% of the
total) (Additional file 1: Table S13), which may be representative of the direct connection to Central
Europe and the Balkan peninsula, respectively.
Quote:On the other hand, it is worth noting
that the trans-Adriatic distribution of the internal branches of J2-M172/M12 was previously interpreted
as a clue of a BA expansion from the Balkans in the Italian area and a link between BA Balkans and BA
Nuragic Sardinia, possibly with peninsular Italian intermediates that were not observed before [19,49].
Interestingly, two out of three of our J2-M12 Picene samples (PN91 and PN101), due to their
phylogenetic position (Additional file 3: Fig. S10) in between the BA Nuragic and the BA Balkan clusters,
could represent the descendants of the aforementioned Italian intermediates
Quote:Despite the diffusion of Near Eastern ancestries seems to be relatively homogeneous and
widespread in the Italian context, in Pesaro necropolis we observe a great genetic variability, towards
both the extremes of the distribution [8,9]. This may be the result of population dynamics occurring in
the Mid-Adriatic area during Late Antiquity. After the fall of the Western Roman Empire, the area was
under control of the Eastern Roman (Byzantine) Empire during the 6th century CE, with the city of
Pesaro representing an important political center of the Duchy of the Pentapolis. It is possible that
these political dynamics additionally influenced the movement of people in the area and, therefore, the
evolution of the genetic pool.
It was only in later periods, starting from the Early Middle Age, that the Italian genetic pool
changed again, with a decrease of Near Eastern ancestry and a new increase of Central/Northern
European one. The main reason for this is probably the massive arrival of people with a Central
European ancestry (like the Longobards) that established the nowadays North-South genetic gradient in
Italy [8,52,53]
https://www.biorxiv.org/content/10.1101/...1.full.pdf
Its a shame they didn't use higher resolution for the yDNA assignments.
Posts: 90
Threads: 4
Joined: Sep 2023
Gender: Male
Nationality: Netherlands
Y-DNA (P): R1b-U152>Z56>Z145
mtDNA (M): W5a2
Country: 
03-20-2024, 10:47 AM
(This post was last modified: 03-20-2024, 11:48 AM by Pylsteen.)
On p18, they suggest that five of the R1b-L23 samples were 'basal' L23, that's not something you often see, if its not P312 or Z2103 maybe PF7589,
well, let's see what the data will tell us. Probably more refinement is possible.
Qrts, Awood, JMcB And 1 others like this post
Posts: 85
Threads: 0
Joined: Sep 2023
Gender: Male
Ethnicity: german/italian
Y-DNA (P): R-YP445 YP729
mtDNA (M): H80
Country: 
03-20-2024, 11:15 AM
(This post was last modified: 03-20-2024, 11:20 AM by alexfritz.)
the impression i get from the PCA plots is that the Picenes (aswell as IA Apulia (Daunians) plot similar as Balkan_IA and this could indicate that a migration did happen across the Adriatic from the Balkan onto the Adriatic coast but there is also an overlap with Etruscans/Latins so prob also a contact zone; they also mention a higher degree of Blondism among the Picenes vs Etruscans/Latins which could further suggest a Balkan migration rather than a diff type of locals
the rest is the same procedure as in the three other papers ie 'population-wide shift' in the principate which the new paper once again stresses how 'population-wide' it actually was
Show Content
SpoilerThe great genetic influence from the Near East in Pesaro (and in general in post-IA Italian populations) is mirrored also in IBD (Additional file 3: Fig. S7) and f4/NNLS analysis in which all the Italian Imperial/Late Antique sites here analyzed cluster together (Additional file 3: Fig. S6).
four of the seven samples do fall into the Imperial/Late-Antiquity space (cf nearby 81.5-220.5 calCE Urbino/Bivio samples); but now we are also dealing with a site from the Byzantine era/area and thus the twe outliers PF1 and PF32 could also be recent arrivals from locs like Antioch and Alexandria (???) while PF15 seems to have had a northward shit; and it must have been a shift rather than an IA relic as the paper mentions that all samples from Pesaro were subject to the prev shift
now it cant really deal with the Longobards (ie Pentapolis) but a minor Ostrogoth site was in the near and the McColl paper assigned R106 and R108 from the Crypta Balbi into the South Scand. cluster, a site i speculated to be Ostrogoth (various hints); nonetheless it also assigned R31 332-419 calCE to the South Scand. cluster and this falls more into the timeperiod of Ricimer and Stilicho (also Odoacer) and their foederati; so smth must have pulled PF15 northward (xLongobards) pre 6th/7th c. ie either the Ostrogoths or prev foederati who now seem to have had an actual presence
Posts: 4
Threads: 0
Joined: Oct 2023
Gender: Male
Nationality: Italian
Y-DNA (P): R1b-U152>L2>Z367>L20
Y-DNA (M): T1a1
Country: 
03-20-2024, 11:37 AM
(This post was last modified: 03-20-2024, 11:47 AM by Gabriel90.)
Dear researchers that work on Roman history and ancient DNA, I have a few questions for you. If Pompeii early imperial samples (79CE) were already significantly more shifted towards the the Near East compared to medieval and modern Southern Italy (let alone Central Italy), why do you think that during high imperial era (especially after 79CE) there was significant genetic influence in Italy from Eastern Mediterranean populations? Wouldn't that process result in a late antiquity and medieval Italy that is more shifted towards the Near East than Pompeii? Contrary to your claims, in late antiquity roman samples we don't have an increase of Near Eastern ancestry, basically early imperial and late imperial samples from Italy and Rome are very similar. If anything, the Late Antiquity roman samples are shifted North, towards Central Europe but especially towards the Western Mediterranean, and not towards the east-med, doesn't that imply that from the first century to the fourth century you had less east-med genetic influence in general? And if the east-med shift is basically completely driven by these "outliers", why does Pompeii had seemingly no locals (with Iron Age profile) and a large amount of intermediate samples (and few or no true Near Eastern samples)? Are we supposed to believe that these intermediate groups did form instantly or maybe we should believe that the process took some generations (and maybe started some centuries earlier)? And most importantly, why do you imply that the time frame for the genetic transition is the imperial era while you have no samples to prove that (from the transition from the republic to the empire), seemingly implying that the change happened overnight? Also remember that Italy had millions of people in the late republic, so you will need millions of people to change to genetic landscape, and also remember that we already had outliers in the previous era (as Hannah Moots pointed out recently on X), many of which from the republican era, even up North in Etruria.
Generally speaking, aren't hypothesis supposed to be proven by data? So why aren't you looking at the transitional period (the middle-late republic)? At no point has anyone really proven that there was a mix of locals (with Iron Age profile) and outliers in Italy in the same time frame, why is that? How can we honestly imply something about the situation without that critical data point? One last thing, often you seem to imply that these outliers are not locals just based on their DNA, whereas isotopes and burial context seem to suggest otherwise, why is that the case?
Finally, are you aware that your studies are used as racist propaganda on social media?
cheshire and Cascio like this post
Posts: 710
Threads: 12
Joined: Sep 2023
(03-20-2024, 11:37 AM)Gabriel90 Wrote: Dear researchers that work on Roman history and ancient DNA, I have a few questions for you. If Pompeii early imperial samples (79CE) were already significantly more shifted towards the the Near East compared to medieval and modern Southern Italy (let alone Central Italy), why do you think that during high imperial era (especially after 79CE) there was significant genetic influence in Italy from Eastern Mediterranean populations? Wouldn't that process result in a late antiquity and medieval Italy that is more shifted towards the Near East than Pompeii? Contrary to your claims, in late antiquity roman samples we don't have an increase of Near Eastern ancestry, basically early imperial and late imperial samples from Italy and Rome are very similar. If anything, the Late Antiquity roman samples are shifted North, towards Central Europe but especially towards the Western Mediterranean, and not towards the east-med, doesn't that imply that from the first century to the fourth century you had less east-med genetic influence in general? And if the east-med shift is basically completely driven by these "outliers", why does Pompeii had seemingly no locals (with Iron Age profile) and a large amount of intermediate samples (and few or no true Near Eastern samples)? Are we supposed to believe that these intermediate groups did form instantly or maybe we should believe that the process took some generations (and maybe started some centuries earlier)?
My guess is that with more data it will be proven that at first coastal, harbor, city and large plantations showed the greatest shift towards the Near East, whereas in later periods the admixture was more widespread throughout the country. The migrants, both free and slaves, landed on specific spots first. Therefore you get at specific sites many completely Near Eastern individuals, whereas in some countryside places it is likely the admixture wasn't arriving at that point in time with the same intensity. Yet it kind of "seeped through" over time, with many of the slaves becoming free, moving around and many free soldiers and others settling down in various places etc.
When looking at Pompeii, and if they are indeed intemediate, we have to assume the process started in this place earlier, some generations before at least. And again it would be logical to assume, that this pattern of freed slaves and settled down migrants which mix with locals started in a place like Pompeii earlier than in some village in the inner countryside.
Posts: 266
Threads: 1
Joined: Oct 2023
03-20-2024, 12:49 PM
(This post was last modified: 03-20-2024, 12:49 PM by corrigendum.)
(03-20-2024, 10:47 AM)Pylsteen Wrote: On p18, they suggest that five of the R1b-L23 samples were 'basal' L23, that's not something you often see, if its not P312 or Z2103 maybe PF7589,
well, let's see what the data will tell us. Probably more refinement is possible.
Quote:Another Picene individual (PN62) belongs to the R1-L23/Z2106 subclade, which has been previously interpreted as a genetic link between Yamnaya, Balkans and Southern Caucasus [19]. Finally, five Picenes and two
Etruscans are placed at the basal portion of the R1-L23 branch, together with other ancient Yamnaya,
Balkan and Southern Caucasic samples (Additional file 2: Fig. S10)
At least one of them is Z2103>Z2106.
Quote:On the other hand, it is worth noting that the trans-Adriatic distribution of the internal branches of J2-M172/M12 was previously interpreted as a clue of a BA expansion from the Balkans in the Italian area and a link between BA Balkans and BA Nuragic Sardinia, possibly with peninsular Italian intermediates that were not observed before [19,49]. Interestingly, two out of three of our J2-M12 Picene samples (PN91 and PN101), due to their phylogenetic position (Additional file 3: Fig. S10) in between the BA Nuragic and the BA Balkan clusters,
could represent the descendants of the aforementioned Italian intermediates[/color][/align]
Quote:From an archaeological perspective, the extensive connections across the two peninsulas throughout the BA and IA are well-characterized. Strong commercial trans-Adriatic routes were already present from the 3rd millennium BCE [7]. During the Early BA the Cetina culture, although rooted in the Dalmatian coast, spread throughout the Adriatic, eventually reaching Sicily, Malta and Western Greece hese contacts persisted throughout the BA [43] and during the IA they were strongly consolidated. Indeed, the extensive presence of shared cultural traits across the two sides of the Adriatic Sea has allowed some authors to describe an “Adriatic koiné” (Adriatic culture) to emphasize this circulation of goods and perhaps individuals [44–46]. Similarly, the possible genetic relationship between Northern/Central Europe and the Middle Adriatic region could be supported by the observed material connections between the Hallstatt culture along the Danube River and Northern-Central Italy, already starting from the Late BA
![[Image: Cetina_Culture_Expansion_Map.png]](https://upload.wikimedia.org/wikipedia/commons/2/22/Cetina_Culture_Expansion_Map.png)
Posts: 27
Threads: 0
Joined: Oct 2023
Country: 
Posts: 4
Threads: 0
Joined: Oct 2023
Gender: Male
Nationality: Italian
Y-DNA (P): R1b-U152>L2>Z367>L20
Y-DNA (M): T1a1
Country: 
03-20-2024, 02:04 PM
(This post was last modified: 03-20-2024, 02:07 PM by Gabriel90.)
(03-20-2024, 12:25 PM)Riverman Wrote: (03-20-2024, 11:37 AM)Gabriel90 Wrote: Dear researchers that work on Roman history and ancient DNA, I have a few questions for you. If Pompeii early imperial samples (79CE) were already significantly more shifted towards the the Near East compared to medieval and modern Southern Italy (let alone Central Italy), why do you think that during high imperial era (especially after 79CE) there was significant genetic influence in Italy from Eastern Mediterranean populations? Wouldn't that process result in a late antiquity and medieval Italy that is more shifted towards the Near East than Pompeii? Contrary to your claims, in late antiquity roman samples we don't have an increase of Near Eastern ancestry, basically early imperial and late imperial samples from Italy and Rome are very similar. If anything, the Late Antiquity roman samples are shifted North, towards Central Europe but especially towards the Western Mediterranean, and not towards the east-med, doesn't that imply that from the first century to the fourth century you had less east-med genetic influence in general? And if the east-med shift is basically completely driven by these "outliers", why does Pompeii had seemingly no locals (with Iron Age profile) and a large amount of intermediate samples (and few or no true Near Eastern samples)? Are we supposed to believe that these intermediate groups did form instantly or maybe we should believe that the process took some generations (and maybe started some centuries earlier)?
My guess is that with more data it will be proven that at first coastal, harbor, city and large plantations showed the greatest shift towards the Near East, whereas in later periods the admixture was more widespread throughout the country. The migrants, both free and slaves, landed on specific spots first. Therefore you get at specific sites many completely Near Eastern individuals, whereas in some countryside places it is likely the admixture wasn't arriving at that point in time with the same intensity. Yet it kind of "seeped through" over time, with many of the slaves becoming free, moving around and many free soldiers and others settling down in various places etc.
When looking at Pompeii, and if they are indeed intemediate, we have to assume the process started in this place earlier, some generations before at least. And again it would be logical to assume, that this pattern of freed slaves and settled down migrants which mix with locals started in a place like Pompeii earlier than in some village in the inner countryside. This is an answer, but not the answer that they gave us in the Moots paper, in the Etruscan paper, in the Pompeii paper and in this one. We already knew that Urbino and Picenum early imperial era samples were less east-med and more Italic than coeval samples from Rome (but only at the level of Southern Italy, so not particularly Italic either). So your theory has some evidence for it. But it doesn't really corroborate the idea that there was a huge influx of foreigners in the first few centuries of the Empire (1th and 2th century), and surely not right before or after the third century crisis.
Reading this paper I think that basically everyone got the idea that during the empire, in a progressive way, especially later on, you had large influx of migrants. Basically early on (in early imperial era) you had a pure Italic-like majority, that was progressively replaced in the middle or late imperial era by foreigners. I think we have enough data to say that this is absolutely bogus. Most of the east-med already came before Pompeii, since most of these samples are adults (so they were born decades before 79CE) and intermediate between Italic\Greek\Gaul and east-med, it's reasonable to believe that especially considering other western influences, Italy as a whole didn't get more east-med during the Empire. So the total balance of migrations during the empire didn't skew east-med. This is a completely opposite conclusion of the published papers about the topic, but IMO it's the correct and obvious interpretation. If more east-med came, than late imperial Rome would have been more East-med than early imperial Rome. It's not the case at all.
Since we have samples from Pompeii, so we solved the cremation bias, and we have samples from diverse areas, some rich and some poor, some rural and some urban, I think that most possible sampling bias have been resolved. I think the samples are representative of the situation and the picture is overall quite clear.
Cascio and pelop like this post
Posts: 7
Threads: 0
Joined: Dec 2023
Gender: Male
Ethnicity: Euro-mix
Y-DNA (P): J-FT86547
mtDNA (M): H1
Country: 
It looks like they took more care refining Mtdna rather then Ydna
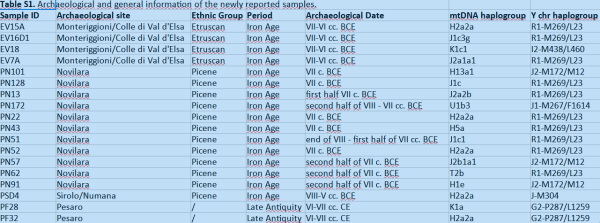
JMcB and Moeca like this post
Posts: 477
Threads: 0
Joined: Oct 2023
Country: 
interesting but I don’t understand the ‘basal L23’ that is described as like a yamnaya when they also distinguish it from Z2103. Doesn’t make sense. Is it not just L23 that they haven’t managed to resolved further?
Manofthehour and Pylsteen like this post
Posts: 51
Threads: 1
Joined: Dec 2023
Country: 
(03-20-2024, 10:35 AM)Riverman Wrote: From the paper:
Quote:Y chromosome data of the Italic IA groups provide additional evidence to these observations,
suggesting that the two scenarios proposed are complementary. Indeed, in the Picenes, two main Y
haplogroups are observed, namely R1-M269/L23 (58% of the total) and J2-M172/M12 (25% of the
total) (Additional file 1: Table S13), which may be representative of the direct connection to Central
Europe and the Balkan peninsula, respectively.
Quote:On the other hand, it is worth noting
that the trans-Adriatic distribution of the internal branches of J2-M172/M12 was previously interpreted
as a clue of a BA expansion from the Balkans in the Italian area and a link between BA Balkans and BA
Nuragic Sardinia, possibly with peninsular Italian intermediates that were not observed before [19,49].
Interestingly, two out of three of our J2-M12 Picene samples (PN91 and PN101), due to their
phylogenetic position (Additional file 3: Fig. S10) in between the BA Nuragic and the BA Balkan clusters,
could represent the descendants of the aforementioned Italian intermediates
https://www.biorxiv.org/content/10.1101/...1.full.pdf
Its a shame they didn't use higher resolution for the yDNA assignments.
Indeed, with the resolution they have (only J-L283 assignement) I don't see how they can claim anything about relative phylogenic placement. Am I missing something ? Or they just doing garbage to have something to say in their paper ?
By IA, in Italy you can nearly expect anything below J-L283 ...
On a phylogenic POV, intermediate samples between Nuragic and Balkanic ones would be (Z2509+, Z585?, Z615-) ... but considering the location of this sample on the Adriatic side, I would expect them to be Z615+ ( and even more likely Z597+, with both Y15058 or Z638 being possible at those dates and location).
While, I'll be the first one happy to see more (Z2509+, Z615-) from Italy, I don't see how, with their resolution, they can claim that those samples are "between" Nuragic and Balkanic ones on a phylogenic POV.
It is a pity that in a published paper we have claims about "BA migrations" using clades that are fully decoupled by ~3100 BCE as a tracer ... It gives the impression of claims made in 10 mins to fill the paper with stuff.
Posts: 27
Threads: 0
Joined: Oct 2023
Country: 
Forgot to add that coverage for most samples is awful.
![[Image: JrlhbeD.png]](https://i.imgur.com/JrlhbeD.png)
Posts: 7
Threads: 0
Joined: Dec 2023
Gender: Male
Ethnicity: Euro-mix
Y-DNA (P): J-FT86547
mtDNA (M): H1
Country: 
03-20-2024, 02:41 PM
(This post was last modified: 03-20-2024, 02:41 PM by Nictus.)
An extract from an other table where there is indeed some more refinement, still not a lot
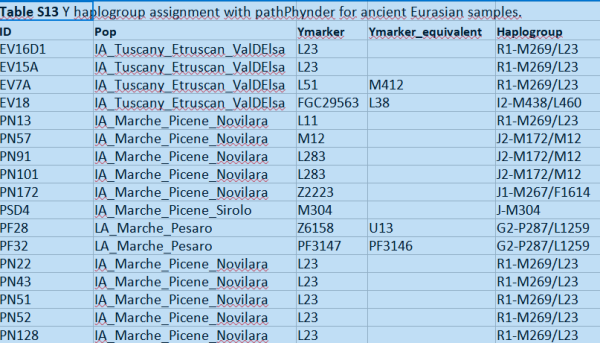
Senhor_Fernandes likes this post
Posts: 13
Threads: 0
Joined: Sep 2023
Gender: Male
Y-DNA (P): J-L283
03-20-2024, 03:36 PM
(This post was last modified: 03-20-2024, 03:52 PM by Trojet.)
In this PDF file ( https://www.biorxiv.org/content/biorxiv/...nload=true) they have a phylogenetic Y tree on "Figure S10" which compares their samples to other aDNA samples. It implies two J2b samples (PN101 and PN91) are parallel to NA20763 and upstream of BA Croatia and BA Montenegro, but downstream of I5723 IA Croatia (J-YP91). So perhaps they are somewhere in between Z600 and Z597.
At least one R1b-M269 is Z2103+.
In any case, it appears the raw data has been uploaded to ENA, so hopefully it will be available soon.
The dataset generated and analyzed during the current study are available in the European
Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena/browser/home) under Accession Number
PRJEB62031.
|








![[Image: Cetina_Culture_Expansion_Map.png]](https://upload.wikimedia.org/wikipedia/commons/2/22/Cetina_Culture_Expansion_Map.png)
![[Image: kRHFnfI.png]](https://i.imgur.com/kRHFnfI.png)
![[Image: EJzIYY9.png]](https://i.imgur.com/EJzIYY9.png)
![[Image: 9v33Yme.png]](https://i.imgur.com/9v33Yme.png)



![[Image: JrlhbeD.png]](https://i.imgur.com/JrlhbeD.png)