Posts: 19
Threads: 0
Joined: Oct 2023
03-04-2024, 05:47 PM
(This post was last modified: 03-04-2024, 05:48 PM by Horatio McCallister.)
I have a hard time buying that Neanderthals really did lose their native Y and Mtdna from introgression from humans. How is it possible that such foundational building blocks of genetic code would be swept out by foreign analogs, why should human Y and MtDna be more optimal against Neanderthal's genetic background compared to their own native Y/Mt? I don't believe the explanation is due to genetic load being higher in Neanderthals, genetic load while very real can be offset salvage mutations which cancel out accumulated deleterious mutations, if Neander Y and mtDNA were really that messed up I find it more likely that they would have native salvage replacements than acquiring new ones from modern humans. Nevermind that I don't think anyone's seriously done a deep dive into Neander Y and MtDNA to see if they were actually significantly messed up to begin with...
What if instead of introgression, Neanderthal and humans are actually just a clade together, with Neanderthal just being sort of hybrid between some proto-human and Denisovan-like groups?
Kaltmeister and AimSmall like this post
Posts: 276
Threads: 2
Joined: Sep 2023
Country: 
(03-04-2024, 05:47 PM)Horatio McCallister Wrote: What if instead of introgression, Neanderthal and humans are actually just a clade together, with Neanderthal just being sort of hybrid between some proto-human and Denisovan-like groups?
That's kind of the same thing, just different proportions. I do think that is indeed the case however.
Posts: 34
Threads: 0
Joined: Oct 2023
Quote:What if instead of introgression, Neanderthal and humans are actually just a clade together, with Neanderthal just being sort of hybrid between some proto-human and Denisovan-like groups?
But that's not how Neandert(h)als are defined. Neanderthals were already present in Europe by 400,000 years ago, when they were still not too divergent from Denisovans, but well after their split.
https://www.mpg.de/10364707/hominins-sima-de-los-huesos
The Neanderthal population of Europe originally had Denisovan-like mtDNA's. The Neanderthal-Modern Human hybrids later spread from SW Asia, replacing all other Neanderthals in both Europe and Asia, hundreds of thousands of years after the split of Denisovans and Neanderthals.
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
(03-04-2024, 05:38 PM)Kale Wrote: (03-04-2024, 04:53 PM)TanTin Wrote: Regarding Spy and Mezmayskaya - I found they have human Y chr by myself. I was playing to project Ychr on PCA and both these Neanderthal projected in between African and non-Africans. Then I found that Spy has many common markers with other A chr individuals. So there are no doubts these 2 Neanderthals are A. I can also give you the list of snips if you need. But you can find them easy, as they are in the file.
You understand that would be huge news, and thus something that would likely have been publicized yes? Consequently skepticism is the reasonable approach. Can you give a list of positive and negative calls for A0-T for example?
The story is more complicated than I expected.
The first match:
rs188292317 - Spy
rs184836128 - both Spy and Mezmaiskaya
rs191856881 - Mezmaiskaya
(45) rs191505182 - both Spy and Mezmaiskaya have it
rs34740450 - Spy <---- This is on the A1b side, however Spy has it !
There are not so many matching snips, still Spy and Mezmaiskaya are on the side of A0 - A haplogroups. I will try to explain that later. The reason: because A-haplogroups start to develop shortly after disconnecting from Neanderthals. And they go to one direction. From the same template or Matrix on the other direction will start to develop the other haplogroups that we know. So that why Spy/ Mezmaiskaya come in the middle.
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
(03-04-2024, 05:47 PM)Horatio McCallister Wrote: I have a hard time buying that Neanderthals really did lose their native Y and Mtdna from introgression from humans. How is it possible that such foundational building blocks of genetic code would be swept out by foreign analogs, why should human Y and MtDna be more optimal against Neanderthal's genetic background compared to their own native Y/Mt? I don't believe the explanation is due to genetic load being higher in Neanderthals, genetic load while very real can be offset salvage mutations which cancel out accumulated deleterious mutations, if Neander Y and mtDNA were really that messed up I find it more likely that they would have native salvage replacements than acquiring new ones from modern humans. Nevermind that I don't think anyone's seriously done a deep dive into Neander Y and MtDNA to see if they were actually significantly messed up to begin with...
What if instead of introgression, Neanderthal and humans are actually just a clade together, with Neanderthal just being sort of hybrid between some proto-human and Denisovan-like groups?
Neanderthals and Denisova are very complicated hybrid.
And they are very different from each other. As an example, I can compare Denisova as 1/2 Chimp + 1/2 Vindija Neanderthal. But it is more complicated than that.
That's not the whole story. The Chimp has 48 chromosomes. (24 dipole)
So if we expect that Neanderthal / Denisova originated from Chimp, then the reduction of the number of chromosomes is a huge obstacle.
Imagine losing 2 chromosomes in short time like 2 million years ?
So this hybridization between Chimp and Humans seems to be almost as catastrophic event. The most surprising is that there were survivals and these survivals continued to survive and change fast. More than that: these Neanderthal and Denisova hybrids created this part of the human ancestors, that we consider originated from Chimp.. Because there is some connection to Chimp, but this connection to Chimp goes via Denisova(Neand.).
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
03-04-2024, 08:18 PM
(This post was last modified: 03-04-2024, 08:50 PM by TanTin.
Edit Reason: Modification
)
To add some more snips in the list (for SPY)
rs112306755 - Spy CTS11573 B~ rs112306755 23163914 21002028 C->T
CTS346 B2~ M8679 rs112994083 6701256 6833215 C->A
M8680 B~ CTS503 rs113481612 6804479 6936438 T->A
Here is the whole list for Spy Neanderhtal:
[1] "rs112994083" "rs113481612" "rs112830147" "rs112750114" "rs113180320" "rs112583133" "rs111518578"
[8] "rs113107044" "rs113244139" "rs113057210" "rs112119188" "rs2032601" "rs112033097" "rs112987230"
[15] "rs113823444" "rs112501714" "rs190560626" "rs111527361" "rs113237637" "rs111309761" "rs111913672"
[22] "rs112396040" "rs111508636" "rs113312178" "rs112891941" "rs113534653" "rs111934740" "rs112400246"
[29] "rs112097234" "rs112272034"
####
Clarification is needed here.
In fact the Spy and Mezmaiskaya are on the opposite side for this list.
They have the opposite snip, same as the other humans who are not from B-haplogroup.
This is part of the initial template or matrix. So then from this template the other haplogroups will appear. The snips value list here will change for the people in hg B ! Other non-B humans will keep the default value for these snips.
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
hg BT
rs189761931 - Both Spy and Mezmaiskaya ( this is a good one!)
rs185180763 --> this is an excellent one !! Spy has it. It is present both in A0, A0a, A1b // No one from OOA will get this one !
rs111335542 - this is also an excellent one !! ( no comments for now, but I will have more to tell )
rs184836128 - already in the first list above
V241 A1 rs184836128 7658712 7790671 C->T
rs111858852
M9000 BT rs111858852 7815370 7947329 T->A
rs112297657 - Both Spy and Mezmaiskaya = "G" <-- Will stay on the side of hg A...
Z40387 BT rs112297657 8042040 8173999 G->A ( this snip will become "A" during "B" haplogroup change , all other humans will keep it as "A")
rs76706607 - same as the previous snip (BT)
And this one is also a very good example:
rs192939307
Spy[ rs192939307 ] = "A"
L413 BT PF1409; V31 rs192939307 6932831 7064790 G->A
PF1409 BT L413; V31 rs192939307 6932831 7064790 G->A
V31 BT L413; PF1409 rs192939307 6932831 7064790 G->A
We don't know if the initial state was A or G.
Chimp allele: G Chimp strand: - Chimp position: chrY:25452653-25452653
In Chimp this snip was "G".. However in Spy it is already "A"..
So you may look at Spy as the very first Human, or half human half Chimp. Those from Hg "A" kept the initial state of "G".
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
Here is how I do my verifications:
![[Image: Haplogroups-A.png]](https://i.postimg.cc/kX5dwyfw/Haplogroups-A.png)
I have already a PCA graph, I just add a color for the specific snips.
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
03-04-2024, 10:13 PM
(This post was last modified: 03-04-2024, 10:22 PM by TanTin.)
And here is another perfect demo. Isn't it amazing ?
rs111683521
M9005 BT rs111683521 7904101 8036060 G->A
Now the most amazing part:
Spy will stay on the side of non_African humans ( haplogroups not A or B)
Mezmaiskaya will stay on the side of A-B Haplogroups.
Where is the Chimp? Chimp is "G" of course, the initial state.
Quote:dbSNP build 150 rs111683521
dbSNP: rs111683521
Position: chrY:7904101-7904101
Band: Yp11.2
Genomic Size: 1
View DNA for this feature (hg19/Human)
Summary: G>A/G (chimp allele displayed first, then '>', then human alleles)
Strand: +
Observed: A/G
Reference allele: A
Chimp allele: G Chimp strand: + Chimp position: chrY_NW_015974592v1_random:606550-606550
Orangutan allele: ?
Macaque allele: ?
- Again, the Spy Neand. will apear as the first human OOA.
![[Image: Haplogroups-A-B.png]](https://i.postimg.cc/L5BSbHW1/Haplogroups-A-B.png)
( the blue color is for missing data or no such snip present, so we just ignore the points in blue. I keep it just as a reference. ).
AimSmall and Quint like this post
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
Another example:
dbSNP build 150 rs74796019
Quote:dbSNP build 150 rs74796019
Summary: G>A/G (chimp allele displayed first, then '>', then human alleles)
Strand: +
Observed: A/G
Reference allele: A
Chimp allele: G Chimp strand: + Chimp position: chrY:15299573-15299573
Orangutan allele: G Orangutan strand: + Orangutan position: chr16:30325486-30325486
Macaque allele: G Macaque strand: - Macaque position: chrUn_NW_014839899v1:5305-5305
Spy[ rs74796019 ] = "A"
Haplogroups A&B are keeping the archaic "G".
There are also other haplogroups keeping the archaic "G". Most of the humans will have the updated snip version "A" .
However the first and the earliest in time appear to be Spy. Spy = "A"
Z40374 BT rs74796019 10033862 10196253 A->G
Posts: 21
Threads: 0
Joined: Oct 2023
Gender: Male
Y-DNA (P): I1-L22
mtDNA (M): T2c1d-a
Spy, that was also the one with the greatest Eurasian "affinity" in G25 before the new samples from this study appeared:
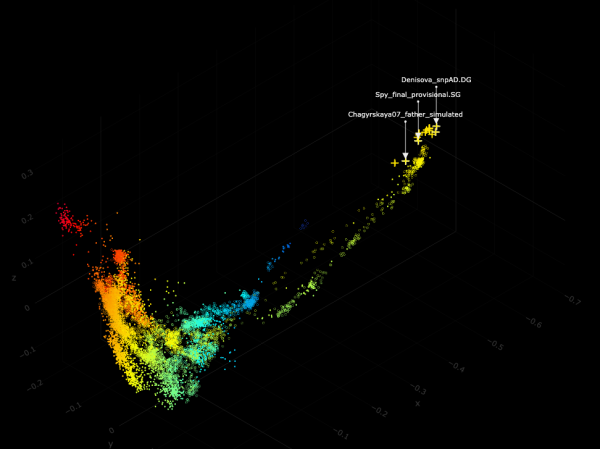 (X: 1, Y: 3, Z: 4)
(X: 1, Y: 3, Z: 4)
Code: Distance difference: ( AC - BC ) ↑
A: Spy_final_provisional.SG
B: Altai_snpAD.DG
C: ↴
-0.04367648 Kamboj:KJ_03
-0.04366371 Pakistani:F069316
-0.04365953 Arain:ARN013
-0.04365612 Pakistani:F073540
-0.04363252 Pashtun_Tarkalani:T-215
-0.04360587 Tajik_Panjshir
-0.04360563 Arain:ARN005
-0.04358377 Pashtun_Yusufzai:Y-216
-0.04357319 Brahui:HGDP00031
-0.04356504 Punjabi_Lahore:HG03767.SG
-0.04356504 Punjabi_Lahore:HG03767.SG
-0.04356353 Pashtun_Tarkalani_Kakazai
-0.04356339 Pakistan_Katelai_IA:I12461
-0.04356335 Khatri:K-69
-0.04355636 Khatri:K-56
-0.04354994 Punjabi_Sikh_India:PanSikh32765
-0.04354013 Kashmiri_Pakistan_o:YPAJK056
-0.04354013 Kashmiri_Pakistan_o:YPAJK056
-0.04353807 Khatri:K-34
-0.04352861 Punjabi_Sikh_India:PanSikh14311
-0.04352648 Punjabi_Lahore:HG02604.SG
-0.04352648 Punjabi_Lahore:HG02604.SG
-0.04352616 Pashtun_Yusufzai:Y-122
-0.04352576 Pashtun_Khattak_Nowshera:PKN_ZA
-0.04352487 Punjabi_Sikh_India:PanSikh28494
Distance difference: ( AC - BC ) ↓
A: Spy_final_provisional.SG
B: Altai_snpAD.DG
C: ↴
0.03984008 Chagyrskaya.SG
0.03791609 Vindija_snpAD.DG
0.03269506 Denisova_snpAD.DG
0.02189159 Goyet_final_provisional.SG
0.02092926 Chimp.REF
0.01654583 Denisova11.SG
0.01226479 Ancestor.REF
0.00886643 Les_Cottes_final_provisional.SG
0.00757514 Mbuti:A_Mbuti-5.DG
0.00733871 Mbuti:S_Mbuti-2.DG
0.00706780 Mbuti
0.00702710 Mbuti:S_Mbuti-1.DG
0.00694033 Mbuti:B_Mbuti-4.DG
0.00639248 Mbuti:S_Mbuti-3.DG
0.00627338 Pygmy:AFP4
0.00559918 Pygmy:AFP8
0.00543924 Pygmy:AFP10
0.00435839 Pygmy:AFP28
0.00425261 Pygmy:AFP29
0.00404787 Pygmy:AFP9
0.00355300 Pygmy:AFP20
0.00349508 Pygmy:AFP25
0.00345691 Pygmy:AFP11
0.00320001 Pygmy:AFP5
0.00316641 Mbuti_(low_res)
Code: Altai_snpAD.DG,-0.664727,0.051792,0.032055,0.067184,0.014772,-0.023985,0.264622,-0.203991,-0.028838,0.056129,0.009256,-0.061295,-0.006987,0.001514,0.010722,-0.015115,0.014212,0.148986,-0.041606,-0.001126,-0.017095,0.001237,-0.002342,-0.007832,-0.010298
Ancestor.REF,-0.667003,0.05687,0.027907,0.055879,0.008925,-0.006972,0.250756,-0.187377,-0.023725,0.040274,0.008282,-0.062794,-0.006838,0.001651,0.008415,-0.011535,0.014864,0.143792,-0.041229,-0.003126,-0.014225,0.001113,-0.001602,-0.005904,-0.010897
Chagyrskaya.SG,-0.663589,0.050776,0.032432,0.068799,0.01508,-0.023706,0.264387,-0.20353,-0.030065,0.057404,0.009581,-0.063244,-0.006838,0.000138,0.009908,-0.014054,0.012386,0.149746,-0.041732,0.001751,-0.017594,0.001484,-0.001109,-0.008073,-0.010658
Chimp.REF,-0.676109,0.057885,0.02753,0.056525,0.009848,-0.00753,0.253106,-0.191992,-0.024134,0.040092,0.008444,-0.061595,-0.005203,0.00234,0.010722,-0.011933,0.015255,0.143792,-0.043869,-0.002501,-0.01435,0.000618,-0.000246,-0.004217,-0.009939
Denisova11.SG,-0.657898,0.051792,0.025267,0.069768,0.017542,-0.02761,0.256162,-0.196607,-0.028633,0.049022,0.008769,-0.057399,-0.002676,-0.000275,0.011129,-0.013524,0.015776,0.13657,-0.037332,0.003752,-0.01722,-0.001484,-0.002218,-0.004458,-0.010538
Denisova_snpAD.DG,-0.674971,0.052808,0.024513,0.071706,0.018773,-0.03514,0.261332,-0.19753,-0.033133,0.046106,0.010555,-0.061595,-0.001933,-0.000826,0.010043,-0.01538,0.015646,0.145692,-0.042235,-0.002251,-0.019216,0.000866,0.000616,-0.003133,-0.015567
Goyet_final_provisional.SG,-0.655621,0.049761,0.030547,0.069768,0.014772,-0.02259,0.257572,-0.20653,-0.029451,0.057769,0.007307,-0.066841,-0.008771,0.000275,0.011536,-0.013922,0.012126,0.145312,-0.040978,-0.001,-0.013975,0.002473,-0.003574,-0.002892,-0.008742
Les_Cottes_final_provisional.SG,-0.651068,0.051792,0.029793,0.067507,0.016003,-0.021753,0.251931,-0.199146,-0.029042,0.056493,0.003573,-0.061745,-0.00773,0,0.012215,-0.013922,0.010822,0.143792,-0.040475,-0.000125,-0.017719,0.001237,-0.002465,-0.005784,-0.007305
Mezmaiskaya1_final_provisional.SG,-0.631718,0.044683,0.030924,0.065246,0.015387,-0.016455,0.246526,-0.193146,-0.027815,0.050662,0.006658,-0.059347,-0.004311,0.004129,0.013708,-0.012861,0.008214,0.145565,-0.037081,0.007629,-0.012977,-0.000371,-0.006655,-0.003133,-0.008981
Mezmaiskaya2_final_provisional.SG,-0.638548,0.049761,0.032809,0.070414,0.016311,-0.021196,0.248876,-0.193838,-0.024134,0.054489,0.01153,-0.062045,-0.011001,0.000138,0.010993,-0.014452,0.0103,0.140878,-0.038966,0.002001,-0.011605,-0.003957,0.001232,-0.004217,-0.011256
Spy_final_provisional.SG,-0.626027,0.052808,0.022627,0.064277,0.010156,-0.017849,0.249816,-0.192684,-0.027202,0.054671,0.006171,-0.063094,-0.008028,-0.00523,0.012893,-0.010077,0.012778,0.149112,-0.042486,0.003377,-0.017095,-0.001607,0,-0.001807,-0.011735
Vindija_snpAD.DG,-0.663589,0.049761,0.030924,0.070091,0.014156,-0.023706,0.263682,-0.203761,-0.02986,0.058133,0.008607,-0.063693,-0.006838,-0.000413,0.010315,-0.015115,0.012256,0.150886,-0.042235,0.000375,-0.014974,0.000371,-0.000863,-0.007109,-0.009819
VindijaG1_final_provisional.SG,-0.637409,0.044683,0.032055,0.067184,0.012925,-0.0251,0.246526,-0.19453,-0.025975,0.050479,0.010393,-0.06714,-0.011596,-0.000826,0.008279,-0.012729,0.01356,0.141384,-0.040349,0.004752,-0.014225,-0.002226,-0.002465,-0.005302,-0.010897
(Thank you to Ajeje Brazorf for making these available on Anthrogenica)
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
I just created a new repository to share some 3D .
Here is one of these PCA - Y chr examples from here.
https://github.com/db-gen/html/blob/main/test1_temp.zip
Please let me know if you are able to download and view it? So I can continue to upload and share more.
AimSmall and Quint like this post
Posts: 348
Threads: 25
Joined: Aug 2023
Gender: Male
Ethnicity: Colonial American
Nationality: American
Y-DNA (P): R1b-U152 >R-FTA96415
Y-DNA (M): I2-P37 > I-BY77146
mtDNA (M): J1b1a1a
mtDNA (P): H66a
Country: 
Download and viewing successful.
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
(03-05-2024, 12:37 AM)AimSmall Wrote: Download and viewing successful.
I am planning to make the 3D as a standard for the next scientific publications.. They will have to adjust. (I mean the scientific community). Because you see what's there. My words and explanation is too poor to describe all the details there.. There is too much.. But for now let go with some simple steps and let advance one step by another.
Posts: 320
Threads: 8
Joined: Oct 2023
Country: 
I will provide another example for SPY..
Here we have a list of 35 snips. And we check other humans if they match to Spy.. If they match, we increase the counter by 1 for each matching snip.
https://github.com/db-gen/html/blob/main...s_list.zip
(if need I can provide the list of the snips). Let me know if you can browse this.
Quint and AimSmall like this post
|







![[Image: Haplogroups-A.png]](https://i.postimg.cc/kX5dwyfw/Haplogroups-A.png)
![[Image: Haplogroups-A-B.png]](https://i.postimg.cc/L5BSbHW1/Haplogroups-A-B.png)