Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
They don't score like Balts, they score like Slavs. Have you seen the distances to modern populations which I posted?
On the other hand, sample R6759 scores like a Balt (which is why I did not post this sample in this thread).
Posts: 737
Threads: 13
Joined: Sep 2023
(03-01-2024, 09:53 PM)Gordius Wrote: (03-01-2024, 09:47 PM)Tomenable Wrote: (03-01-2024, 09:45 PM)Gordius Wrote: (03-01-2024, 04:21 PM)Tomenable Wrote: What about ancient samples from Himera and Viminacium? You will not add them?
Are they Slavs?
I think so, because they have Slavic-like autosomal results (see above) and also typically Slavic Y-DNA haplogroups.
They have dominating component that we call "Balto-Slavic drift", but this does not mean that they were Slavs. They could be Balts, or they could be a separate branch (both Indo-European and non-Indo-European)
Same thought. Especially since their burial context is not that clear also.
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
03-01-2024, 10:09 PM
(This post was last modified: 03-01-2024, 10:12 PM by Tomenable.)
This is what they score in K36 - their levels of East Central Euro and Eastern Euro are too low for Balts:
![[Image: YLrJ9pZ.png]](https://i.imgur.com/YLrJ9pZ.png)
You can check them with this tool if you want:
https://www.allelocator.ovh/Taux%20de%20Similitude.htm
https://www.exploreyourdna.com/similitude.htm
Posts: 121
Threads: 5
Joined: Sep 2023
(03-01-2024, 09:58 PM)Tomenable Wrote: They don't score like Balts, they score like Slavs. Have you seen the distances to modern populations which I posted?
On the other hand, sample R6759 scores like a Balt (which is why I did not post this sample in this thread).
They score like Slavs because they have more "mediterranean" components than Balts (but some Lithuanians also have the same). For example if a man has 3 granparents Lithuanians and one gransparent Italian - he would score like a Slav.
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
03-01-2024, 10:14 PM
(This post was last modified: 03-01-2024, 10:15 PM by Tomenable.)
For comparison R6759 from Viminacium (this one indeed scores like a Balt):
R6759
Amerindian 0
Arabian 0
Armenian 0
Basque 0
Central African 0
Central Euro 4.11
East African 0
East Asian 0
East Balkan 4.85
East Central Asian 0
East Central Euro 37.35
East Med 0
Eastern Euro 28.93
Fennoscandian 16.53
French 0
Iberian 0
Indo-Chinese 0
Italian 0
Malayan 0
Near Eastern 0
North African 0
North Atlantic 2.68
North Caucasian 0
North Sea 0.74
Northeast African 0
Oceanian 0
Omotic 0
Pygmy 0.06
Siberian 0
South Asian 0
South Central Asian 0
South Chinese 0
Volga-Ural 4.75
West African 0
West Caucasian 0
West Med 0
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
03-01-2024, 10:19 PM
(This post was last modified: 03-01-2024, 10:20 PM by Tomenable.)
(03-01-2024, 10:13 PM)Gordius Wrote: (03-01-2024, 09:58 PM)Tomenable Wrote: They don't score like Balts, they score like Slavs. Have you seen the distances to modern populations which I posted?
On the other hand, sample R6759 scores like a Balt (which is why I did not post this sample in this thread).
They score like Slavs because they have more "mediterranean" components than Balts (but some Lithuanians also have the same). For example if a man has 3 granparents Lithuanians and one gransparent Italian - he would score like a Slav.
North Sea and North Atlantic are not "mediterranean" components, and this appears to be the main difference:
(French is also not "mediterranean" because French in K36 is based mainly on samples from Bretagne, IIRC)
![[Image: dfm5g4U.png]](https://i.imgur.com/dfm5g4U.png)
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
03-01-2024, 10:26 PM
(This post was last modified: 03-01-2024, 10:28 PM by Tomenable.)
Quote:(but some Lithuanians also have the same)
These are either Lithuanians with recent Slavic admixture, or just Slavs from Lithuania (Poles, Belarusians, Russians).
Indigenous Lithuanians without Slavic admixture don't score less than 30% of East Central Euro component in K36.
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
03-01-2024, 10:47 PM
(This post was last modified: 03-01-2024, 11:01 PM by Tomenable.)
Viminacium Slavs also share IBD segments with Medieval Slavic samples according to Vyazov et al. study.
Is it not enough evidence that they were Slavs (combined with autosomal similarity and Y-DNA lineages)?
Posts: 121
Threads: 5
Joined: Sep 2023
(03-01-2024, 10:47 PM)Tomenable Wrote: Viminacium Slavs also share IBD segments with Medieval Slavic samples according to Vyazov et al. study.
Is it not enough evidence that they were Slavs (combined with autosomal similarity and Y-DNA lineages)?
Can you give the link?
Posts: 619
Threads: 63
Joined: Sep 2023
Gender: Male
Ethnicity: Berber
Nationality: North Africa
Y-DNA (P): E-V257(×M81)
Y-DNA (M): V
mtDNA (P): L2b
(10-21-2023, 09:21 PM)Tora_sama Wrote: https://www.google.com/maps/d/viewer?mid...375046&z=5
Who else had autosomes in G25, then there they were checked for proximity to the general Slavic populations.
With the appearance of new samples, the map will be update. To simplify the marking of haplogroups, the same colors are used as in FTDNA, if applications with the texture of the haplogroup were found in one place, then the color of the predominant one was indicated. Places with MT haplogroups only are indicated by color.
If you want to add a sample, then you need to:
1)Place
2)Haplogroups
3)Dating
4)Link to the source
Your comments and suggestions will be accepted, so do not hesitate to write.
Hello , do you can added sample KRA005 its seems to its slavic ancestry
Y-DNA:E-S3003*?
mtDNA:H23*
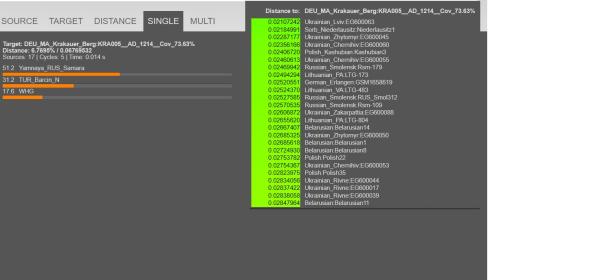
Quote:DEU_MA_Krakauer_Berg:KRA005__AD_1214__Cov_73.63%,0.127482,0.13405,0.069013,0.068799,0.049548,0.021753,0.009635,0.015461,-0.006545,-0.021139,-0.004709,-0.009442,0.010406,0.02257,-0.006515,-0.004508,0.009909,-0.001774,0.006411,0.006753,-0.008235,-0.002968,0.006655,-0.00482,0.000239
https://www.nature.com/articles/s41598-020-75163-w
Tora_sama likes this post
Target: CapsianWGS_scaled
Distance: 1.2510% / 0.01251049
37.2 Iberomaurusian
36.8 Early_European_Farmer
12.8 Early_Levantine_Farmer
8.0 Steppe_Pastoralist
4.8 SSA
0.4 Iran_Neolithic
FTDNA : 91% North Africa +<2% Bedouin + <2 Southern-Levantinfo + <1 Sephardic Jewish + 3% Malta + 3% Iberian Peninsula
23andME : 100% North Africa
WGS ( Y-DNA and mtDNA)
Y-DNA: E-A30032< A30480 ~1610 CE
mtDNA: V25b 800CE ? ( age mtDNA not accurate )
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
(03-02-2024, 09:11 AM)Gordius Wrote: (03-01-2024, 10:47 PM)Tomenable Wrote: Viminacium Slavs also share IBD segments with Medieval Slavic samples according to Vyazov et al. study.
Is it not enough evidence that they were Slavs (combined with autosomal similarity and Y-DNA lineages)?
Can you give the link?
"Slavs cluster" refers to the Slavic IBD cluster, and as you can see Viminacium is one of the dots:
https://i.postimg.cc/WVQsX9rR/Screenshot...3-Zoom.jpg
Posts: 40
Threads: 3
Joined: Oct 2023
(03-01-2024, 10:47 PM)Tomenable Wrote: Viminacium Slavs also share IBD segments with Medieval Slavic samples according to Vyazov et al. study.
Is it not enough evidence that they were Slavs (combined with autosomal similarity and Y-DNA lineages)?
Just to understand power of ibd methodology in telling Balt from Slav.
Would Lithuania IA (Marvele) share IBD with Medieval Slavic samples or not?
Would modern Latvians share IBD with Medieval Slavic or not?
Would BA Baltics (EST, LVA) share IBD or not?
Posts: 121
Threads: 5
Joined: Sep 2023
(03-02-2024, 10:21 PM)Tomenable Wrote: (03-02-2024, 09:11 AM)Gordius Wrote: (03-01-2024, 10:47 PM)Tomenable Wrote: Viminacium Slavs also share IBD segments with Medieval Slavic samples according to Vyazov et al. study.
Is it not enough evidence that they were Slavs (combined with autosomal similarity and Y-DNA lineages)?
Can you give the link?
"Slavs cluster" refers to the Slavic IBD cluster, and as you can see Viminacium is one of the dots:
https://i.postimg.cc/WVQsX9rR/Screenshot...3-Zoom.jpg
What study is this?
Posts: 411
Threads: 15
Joined: Oct 2023
Gender: Male
Nationality: Polish
Y-DNA (P): R1b-BY194358
Y-DNA (M): R1a-FTD42626
mtDNA (M): W6a
Country: 
(03-03-2024, 06:33 PM)Gordius Wrote: What study is this?
The study has not yet been published, but it will be. The screenshot is from a presentation, but I cannot post more screenshots from it.
Posts: 11
Threads: 1
Joined: Oct 2023
Gender: Male
Nationality: Russian
Y-DNA (P): E-Y184711
mtDNA (M): H1b2g
(03-01-2024, 04:21 PM)Tomenable Wrote: So there are still only two Slavic samples from the oldest period 6th - 9th centuries?
What about ancient samples from Himera and Viminacium? You will not add them?
There is no point in adding samples earlier than the 6th century, since actually from the 6th century the Slavs began to be called and be Slavs.
Vinitharya likes this post
|






![[Image: YLrJ9pZ.png]](https://i.imgur.com/YLrJ9pZ.png)
![[Image: dfm5g4U.png]](https://i.imgur.com/dfm5g4U.png)